Covalent API#
The following API documentation describes how to use Covalent.
The Covalent Server manages workflow dispatch, orchestration, and metadata
Quantum Electrons are used to customize and track quantum circuit execution
Quantum Clusters are used to distribute Quantum Electrons across multiple quantum backends.
Local Executor is used to execute electrons locally
File and Data Transfer is used to queue remote or local file transfer operations prior or post electron execution.
File Transfer Strategies are used to perform download/upload/copy operations over various protocols.
Automate Repetitive Tasks with Triggers are used to execute a workflow triggered by a specific type of event
Dask Executor is used to execute electrons in a Dask cluster
Dependencies are used to specify any kind of electron dependency
Pip Dependencies are used to specify PyPI packages that are required to run an electron
Bash Dependencies are used to specify optional pre-execution shell commands for an electron
Call Dependencies are used to specify functions or dependencies that are called in an electron’s execution environment
Results is used for collecting and manipulating results
Dispatcher is used for dispatching workflows and stopping triggered dispatches
The dispatcher_server_api is used for interfacing with the Covalent server
Covalent Server#
A Covalent server must be running in order to dispatch workflows. The Covalent CLI provides various utilities for starting, stopping, and managing a Covalent server. For more information, see:
covalent --help
The Covalent SDK also includes a Python interface for starting and stopping the Covalent server.
Electron#
- class covalent._workflow.electron.Electron(function, node_id=None, metadata=None, task_group_id=None, packing_tasks=False)[source]#
An electron (or task) object that is a modular component of a work flow and is returned by
electron.- function#
Function to be executed.
- node_id#
Node id of the electron.
- metadata#
Metadata to be used for the function execution.
- kwargs#
Keyword arguments if any.
- task_group_id#
the group to which the task be assigned when it is bound to a graph node. If unset, the group id will default to node id.
- packing_tasks#
Flag to indicate whether task packing is enabled.
Attributes:
Get transportable electron object and metadata.
Methods:
connect_node_with_others(node_id, …)Adds a node along with connecting edges for all the arguments to the electron.
get_metadata(name)Get value of the metadata of given name.
get_op_function(operand_1, operand_2, op)Function to handle binary operations with electrons as operands.
set_metadata(name, value)Function to add/edit metadata of given name and value to electron’s metadata.
wait_for(electrons)Waits for the given electrons to complete before executing this one.
- property as_transportable_dict: Dict#
Get transportable electron object and metadata.
- Return type
Dict
- connect_node_with_others(node_id, param_name, param_value, param_type, arg_index, transport_graph)[source]#
Adds a node along with connecting edges for all the arguments to the electron.
- Parameters
node_id (
int) – Node number of the electronparam_name (
str) – Name of the parameterparam_value (
Union[Any,ForwardRef]) – Value of the parameterparam_type (
str) – Type of parameter, positional or keywordtransport_graph (_TransportGraph) – Transport graph of the lattice
- Returns
None
- get_metadata(name)[source]#
Get value of the metadata of given name.
- Parameters
name (
str) – Name of the metadata whose value is needed.- Returns
Value of the metadata of given name.
- Return type
value
- Raises
KeyError – If metadata of given name is not present.
- get_op_function(operand_1, operand_2, op)[source]#
Function to handle binary operations with electrons as operands. This will not execute the operation but rather create another electron which will be postponed to be executed according to the default electron configuration/metadata.
This also makes sure that if these operations are being performed outside of a lattice, then they are performed as is.
- Parameters
- Returns
- Electron object corresponding to the operation execution.
Behaves as a normal function call if outside a lattice.
- Return type
electron
- set_metadata(name, value)[source]#
Function to add/edit metadata of given name and value to electron’s metadata.
- Parameters
name (
str) – Name of the metadata to be added/edited.value (
Any) – Value of the metadata to be added/edited.
- Return type
None- Returns
None
Lattice#
- class covalent._workflow.lattice.Lattice(workflow_function, transport_graph=None)[source]#
A lattice workflow object that holds the work flow graph and is returned by
latticedecorator.- workflow_function#
The workflow function that is decorated by
latticedecorator.
- transport_graph#
The transport graph which will be the basis on how the workflow is executed.
- metadata#
Dictionary of metadata of the lattice.
- post_processing#
Boolean to indicate if the lattice is in post processing mode or not.
- kwargs#
Keyword arguments passed to the workflow function.
- electron_outputs#
Dictionary of electron outputs received after workflow execution.
Methods:
build_graph(*args, **kwargs)Builds the transport graph for the lattice by executing the workflow function which will trigger the call of all underlying electrons and they will get added to the transport graph for later execution.
dispatch(*args, **kwargs)DEPRECATED: Function to dispatch workflows.
dispatch_sync(*args, **kwargs)DEPRECATED: Function to dispatch workflows synchronously by waiting for the result too.
draw(*args, **kwargs)Generate lattice graph and display in UI taking into account passed in arguments.
get_metadata(name)Get value of the metadata of given name.
set_metadata(name, value)Function to add/edit metadata of given name and value to lattice’s metadata.
- build_graph(*args, **kwargs)[source]#
Builds the transport graph for the lattice by executing the workflow function which will trigger the call of all underlying electrons and they will get added to the transport graph for later execution.
Also redirects any print statements inside the lattice function to null and ignores any exceptions caused while executing the function.
GRAPH WILL NOT BE BUILT AFTER AN EXCEPTION HAS OCCURRED.
- Parameters
*args – Positional arguments to be passed to the workflow function.
**kwargs – Keyword arguments to be passed to the workflow function.
- Return type
None- Returns
None
- dispatch(*args, **kwargs)[source]#
DEPRECATED: Function to dispatch workflows.
- Parameters
*args – Positional arguments for the workflow
**kwargs – Keyword arguments for the workflow
- Return type
str- Returns
Dispatch id assigned to job
- dispatch_sync(*args, **kwargs)[source]#
DEPRECATED: Function to dispatch workflows synchronously by waiting for the result too.
- Parameters
*args – Positional arguments for the workflow
**kwargs – Keyword arguments for the workflow
- Return type
- Returns
Result of workflow execution
- draw(*args, **kwargs)[source]#
Generate lattice graph and display in UI taking into account passed in arguments.
- Parameters
*args – Positional arguments to be passed to build the graph.
**kwargs – Keyword arguments to be passed to build the graph.
- Return type
None- Returns
None
Quantum Electrons#
Quantum Clusters#
Local Executor#
Executing tasks (electrons) directly on the local machine
File Transfer#
File Transfer from (source) and to (destination) local or remote files prior/post electron execution. Instances are provided to files keyword argument in an electron decorator.
- class covalent._file_transfer.file.File(filepath=None, is_remote=False, is_dir=False, include_folder=False)[source]#
File class to store components of provided URI including scheme (s3://, file://, ect.) determine if the file is remote, and acts a facade to facilitate filesystem operations.
- filepath#
File path corresponding to the file.
- is_remote#
Flag determining if file is remote (override). Default is resolved automatically from file scheme.
- is_dir#
Flag determining if file is a directory (override). Default is determined if file uri contains trailing slash.
- include_folder#
Flag that determines if the folder should be included in the file transfer, if False only contents of folder are transfered.
- class covalent._file_transfer.folder.Folder(filepath=None, is_remote=False, is_dir=True, include_folder=False)[source]#
Folder class to store components of provided URI including scheme (s3://, file://, ect.), determine if the file is remote, and act as facade to facilitate filesystem operations. Folder is a child of the File class which sets is_dir flag to True.
- include_folder#
Flag that determines if the folder should be included in the file transfer, if False only contents of folder are transfered.
- class covalent._file_transfer.file_transfer.FileTransfer(from_file=None, to_file=None, order=<Order.BEFORE: 'before'>, strategy=None)[source]#
FileTransfer object class that takes two File objects or filepaths (from, to) and a File Transfer Strategy to perform remote or local file transfer operations.
- from_file#
Filepath or File object corresponding to the source file.
- to_file#
Filepath or File object corresponding to the destination file.
- order#
Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution.
- strategy#
Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.
- covalent._file_transfer.file_transfer.TransferFromRemote(from_filepath, to_filepath=None, strategy=None, order=<Order.BEFORE: 'before'>)[source]#
Factory for creating a FileTransfer instance where from_filepath is implicitly created as a remote File Object, and the order (Order.BEFORE) is set so that this file transfer will occur prior to electron execution.
- Parameters
from_filepath (
str) – File path corresponding to remote file (source).to_filepath (
Optional[str]) – File path corresponding to local file (destination)strategy (
Optional[FileTransferStrategy]) – Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.order (
Order) – Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution - default is BEFORE
- Return type
- Returns
FileTransfer instance with implicit Order.BEFORE enum set and from (source) file marked as remote
- covalent._file_transfer.file_transfer.TransferToRemote(to_filepath, from_filepath=None, strategy=None, order=<Order.AFTER: 'after'>)[source]#
Factory for creating a FileTransfer instance where to_filepath is implicitly created as a remote File Object, and the order (Order.AFTER) is set so that this file transfer will occur post electron execution.
- Parameters
to_filepath (
str) – File path corresponding to remote file (destination)from_filepath (
Optional[str]) – File path corresponding to local file (source).strategy (
Optional[FileTransferStrategy]) – Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.order (
Order) – Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution - default is AFTER
- Return type
- Returns
FileTransfer instance with implicit Order.AFTER enum set and to (destination) file marked as remote
File Transfer Strategies#
A set of classes with a shared interface to perform copy, download, and upload operations given two (source & destination) File objects that support various protocols.
- class covalent._file_transfer.strategies.transfer_strategy_base.FileTransferStrategy[source]#
Base FileTransferStrategy class that defines the interface for file transfer strategies exposing common methods for performing copy, download, and upload operations.
- class covalent._file_transfer.strategies.rsync_strategy.Rsync(user='', host='', private_key_path=None)[source]#
Implements Base FileTransferStrategy class to use rsync to move files to and from remote or local filesystems. Rsync via ssh is used if one of the provided files is marked as remote.
- user#
(optional) Determine user to specify for remote host if using rsync with ssh
- host#
(optional) Determine what host to connect to if using rsync with ssh
- private_key_path#
(optional) Filepath for ssh private key to use if using rsync with ssh
Triggers#
Execute a workflow triggered by a specific type of event
Classes:
|
Base class to be subclassed by any custom defined trigger. |
|
Database trigger which can read for changes in a database and trigger workflows based on record changes. |
|
Directory or File based trigger which watches for events in said file/dir and performs a trigger action whenever they happen. |
|
SQLite based Trigger which can read for changes in a SQLite database and trigger workflows based on that. |
|
Performs a trigger action every time_gap seconds. |
- class covalent.triggers.BaseTrigger(lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
objectBase class to be subclassed by any custom defined trigger. Implements all the necessary methods used for interacting with dispatches, including getting their statuses and performing a redispatch of them whenever the trigger gets triggered.
- Parameters
lattice_dispatch_id (
Optional[str]) – Dispatch ID of the worfklow which has to be redispatched in case this trigger gets triggereddispatcher_addr (
Optional[str]) – Address of dispatcher server used to retrieve info about or redispatch any dispatchestriggers_server_addr (
Optional[str]) – Address of the Triggers server (if there is any) to register this trigger to, uses the dispatcher’s address by default
- self.lattice_dispatch_id#
Dispatch ID of the worfklow which has to be redispatched in case this trigger gets triggered
- self.dispatcher_addr#
Address of dispatcher server used to retrieve info about or redispatch any dispatches
- self.triggers_server_addr#
Address of the Triggers server (if there is any) to register this trigger to, uses the dispatcher’s address by default
- self.new_dispatch_ids#
List of all the newly created dispatch ids from performing redispatch
- self.observe_blocks#
Boolean to indicate whether the self.observe method is a blocking call
- self.event_loop#
Event loop to be used if directly calling dispatcher’s functions instead of the REST APIs
- self.use_internal_funcs#
Boolean indicating whether to use dispatcher’s functions directly instead of through API calls
- self.stop_flag#
To handle stopping mechanism in a thread safe manner in case self.observe() is a blocking call (e.g. see TimeTrigger)
Methods:
observe()Start observing for any change which can be used to trigger this trigger.
register()Register this trigger to the Triggers server and start observing.
stop()Stop observing for changes.
to_dict()Return a dictionary representation of this trigger which can later be used to regenerate it.
trigger()Trigger this trigger and perform a redispatch of the connected dispatch id’s workflow.
- abstract observe()[source]#
Start observing for any change which can be used to trigger this trigger. To be implemented by the subclass.
- register()[source]#
Register this trigger to the Triggers server and start observing.
- Return type
None
- class covalent.triggers.DatabaseTrigger(db_path, table_name, poll_interval=1, where_clauses=None, trigger_after_n=1, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerDatabase trigger which can read for changes in a database and trigger workflows based on record changes.
- Parameters
db_path (
str) – Connection string for the databasetable_name (
str) – Name of the table to observepoll_interval (
int) – Time in seconds to wait for before reading the database againwhere_clauses (
Optional[List[str]]) – List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the databasetrigger_after_n (
int) – Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.lattice_dispatch_id (
Optional[str]) – Lattice dispatch id of the workflow to be triggereddispatcher_addr (
Optional[str]) – Address of the dispatcher servertriggers_server_addr (
Optional[str]) – Address of the triggers server
- self.db_path#
Connection string for the database
- self.table_name#
Name of the table to observe
- self.poll_interval#
Time in seconds to wait for before reading the database again
- self.where_clauses#
List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the database
- self.trigger_after_n#
Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action as long as where conditions are met or until stop has being called
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
- class covalent.triggers.DirTrigger(dir_path, event_names, batch_size=1, recursive=False, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerDirectory or File based trigger which watches for events in said file/dir and performs a trigger action whenever they happen.
- Parameters
dir_path – Path to the file/dir which is to be observed for events
event_names – List of event names on which to perform the trigger action. Possible options can be a subset of: [“created”, “deleted”, “modified”, “moved”, “closed”].
batch_size (
int) – The number of changes to wait for before performing the trigger action, default is 1.recursive (
bool) – Whether to recursively watch the directory, default is False.
- self.dir_path#
Path to the file/dir which is to be observed for events
- self.event_names#
List of event names on which to perform the trigger action. Possible options can be a subset of: [“created”, “deleted”, “modified”, “moved”, “closed”]
- self.batch_size#
The number of events to wait for before performing the trigger action, default is 1.
- self.recursive#
Whether to recursively watch the directory, default is False.
- self.n_changes#
Number of events since last trigger action. Whenever self.n_changes == self.batch_size a trigger action happens.
Methods:
Dynamically attaches and overrides the “on_*” methods to the handler depending on which ones are requested by the user.
observe()Start observing the file/dir for any possible events among the ones mentioned in self.event_names.
stop()Stop observing the file or directory for changes.
- attach_methods_to_handler()[source]#
Dynamically attaches and overrides the “on_*” methods to the handler depending on which ones are requested by the user.
- Parameters
event_names – List of event names upon which to perform a trigger action
- Return type
None
- class covalent.triggers.SQLiteTrigger(db_path, table_name, poll_interval=1, where_clauses=None, trigger_after_n=1, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerSQLite based Trigger which can read for changes in a SQLite database and trigger workflows based on that.
- Parameters
db_path (
str) – Absolute path to the database filetable_name (
str) – Name of the table to observepoll_interval (
int) – Time in seconds to wait for before reading the database againwhere_clauses (
Optional[List[str]]) – List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the databasetrigger_after_n (
int) – Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.lattice_dispatch_id (
Optional[str]) – Lattice dispatch id of the workflow to be triggereddispatcher_addr (
Optional[str]) – Address of the dispatcher servertriggers_server_addr (
Optional[str]) – Address of the triggers server
- self.db_path#
Absolute path to the database file
- self.table_name#
Name of the table to observe
- self.poll_interval#
Time in seconds to wait for before reading the database again
- self.where_clauses#
List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the database
- self.trigger_after_n#
Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action as long as where conditions are met or until stop has being called
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
- class covalent.triggers.TimeTrigger(time_gap, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerPerforms a trigger action every time_gap seconds.
- Parameters
time_gap (
int) – Amount of seconds to wait before doing a trigger action
- self.time_gap#
Amount of seconds to wait before doing a trigger action
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action every self.time_gap seconds until stop condition has been met.
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
Dask Executor#
Executing tasks (electrons) in a Dask cluster
Dependencies#
Generic dependencies for an electron
- class covalent._workflow.deps.Deps(apply_fn=None, apply_args=[], apply_kwargs={}, *, retval_keyword='')[source]#
Generic dependency class used in specifying any kind of dependency for an electron.
- apply_fn#
function to be executed in the backend environment
- apply_args#
list of arguments to be applied in the backend environment
- apply_kwargs#
dictionary of keyword arguments to be applied in the backend environment
Methods:
apply()Encapsulates the exact function and args/kwargs to be executed in the backend environment.
- apply()[source]#
Encapsulates the exact function and args/kwargs to be executed in the backend environment.
- Parameters
None –
- Return type
Tuple[TransportableObject,TransportableObject,TransportableObject,str]- Returns
A tuple of transportable objects containing the function and optional args/kwargs
Pip Dependencies#
PyPI packages to be installed before executing an electron
- class covalent._workflow.depspip.DepsPip(packages=[], reqs_path='')[source]#
PyPI packages to be installed before executing an electron
A specification of Pip packages to be installed
- packages#
A list of PyPI packages to install
- reqs_path#
Path to requirements.txt (overrides packages)
These packages are installed in an electron’s execution environment just before the electron is run.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Bash Dependencies#
Shell commands to run before an electron
- class covalent._workflow.depsbash.DepsBash(commands=[])[source]#
Shell commands to run before an electron
Deps class to encapsulate Bash dependencies for an electron.
The specified commands will be executed as subprocesses in the same environment as the electron.
- commands#
A list of bash commands to execute before the electron runs.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Call Dependencies#
Functions, shell commands, PyPI packages, and other types of dependencies to be called in an electron’s execution environment
- class covalent._workflow.depscall.DepsCall(func=None, args=None, kwargs=None, *, retval_keyword='', override_reserved_retval_keys=False)[source]#
Functions, shell commands, PyPI packages, and other types of dependencies to be called in an electron’s execution environment
Deps class to encapsulate python functions to be called in the same execution environment as the electron.
- func#
A callable
- args#
args list
- kwargs#
kwargs dict
- retval_keyword#
An optional string referencing the return value of func.
If retval_keyword is specified, the return value of func will be passed during workflow execution as an argument to the electron corresponding to the parameter of the same name.
Notes
Electron parameters to be injected during execution must have default parameter values.
It is the user’s responsibility to ensure that retval_keyword is actually a parameter of the electron. Unexpected behavior may occur otherwise.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Index#
Here is an alphabetical index.
Contents#
Workflow Components#
Task Helpers#
Executors#
Dispatch Infrastructure#
Covalent CLI Tool#
API#
Workflow Components#
Electron#
- class covalent._workflow.electron.Electron(function, node_id=None, metadata=None, task_group_id=None, packing_tasks=False)[source]#
An electron (or task) object that is a modular component of a work flow and is returned by
electron.- function#
Function to be executed.
- node_id#
Node id of the electron.
- metadata#
Metadata to be used for the function execution.
- kwargs#
Keyword arguments if any.
- task_group_id#
the group to which the task be assigned when it is bound to a graph node. If unset, the group id will default to node id.
- packing_tasks#
Flag to indicate whether task packing is enabled.
Attributes:
Get transportable electron object and metadata.
Methods:
connect_node_with_others(node_id, …)Adds a node along with connecting edges for all the arguments to the electron.
get_metadata(name)Get value of the metadata of given name.
get_op_function(operand_1, operand_2, op)Function to handle binary operations with electrons as operands.
set_metadata(name, value)Function to add/edit metadata of given name and value to electron’s metadata.
wait_for(electrons)Waits for the given electrons to complete before executing this one.
- property as_transportable_dict: Dict#
Get transportable electron object and metadata.
- Return type
Dict
- connect_node_with_others(node_id, param_name, param_value, param_type, arg_index, transport_graph)[source]#
Adds a node along with connecting edges for all the arguments to the electron.
- Parameters
node_id (
int) – Node number of the electronparam_name (
str) – Name of the parameterparam_value (
Union[Any,ForwardRef]) – Value of the parameterparam_type (
str) – Type of parameter, positional or keywordtransport_graph (_TransportGraph) – Transport graph of the lattice
- Returns
None
- get_metadata(name)[source]#
Get value of the metadata of given name.
- Parameters
name (
str) – Name of the metadata whose value is needed.- Returns
Value of the metadata of given name.
- Return type
value
- Raises
KeyError – If metadata of given name is not present.
- get_op_function(operand_1, operand_2, op)[source]#
Function to handle binary operations with electrons as operands. This will not execute the operation but rather create another electron which will be postponed to be executed according to the default electron configuration/metadata.
This also makes sure that if these operations are being performed outside of a lattice, then they are performed as is.
- Parameters
- Returns
- Electron object corresponding to the operation execution.
Behaves as a normal function call if outside a lattice.
- Return type
electron
- set_metadata(name, value)[source]#
Function to add/edit metadata of given name and value to electron’s metadata.
- Parameters
name (
str) – Name of the metadata to be added/edited.value (
Any) – Value of the metadata to be added/edited.
- Return type
None- Returns
None
Lattice#
- class covalent._workflow.lattice.Lattice(workflow_function, transport_graph=None)[source]#
A lattice workflow object that holds the work flow graph and is returned by
latticedecorator.- workflow_function#
The workflow function that is decorated by
latticedecorator.
- transport_graph#
The transport graph which will be the basis on how the workflow is executed.
- metadata#
Dictionary of metadata of the lattice.
- post_processing#
Boolean to indicate if the lattice is in post processing mode or not.
- kwargs#
Keyword arguments passed to the workflow function.
- electron_outputs#
Dictionary of electron outputs received after workflow execution.
Methods:
build_graph(*args, **kwargs)Builds the transport graph for the lattice by executing the workflow function which will trigger the call of all underlying electrons and they will get added to the transport graph for later execution.
dispatch(*args, **kwargs)DEPRECATED: Function to dispatch workflows.
dispatch_sync(*args, **kwargs)DEPRECATED: Function to dispatch workflows synchronously by waiting for the result too.
draw(*args, **kwargs)Generate lattice graph and display in UI taking into account passed in arguments.
get_metadata(name)Get value of the metadata of given name.
set_metadata(name, value)Function to add/edit metadata of given name and value to lattice’s metadata.
- build_graph(*args, **kwargs)[source]#
Builds the transport graph for the lattice by executing the workflow function which will trigger the call of all underlying electrons and they will get added to the transport graph for later execution.
Also redirects any print statements inside the lattice function to null and ignores any exceptions caused while executing the function.
GRAPH WILL NOT BE BUILT AFTER AN EXCEPTION HAS OCCURRED.
- Parameters
*args – Positional arguments to be passed to the workflow function.
**kwargs – Keyword arguments to be passed to the workflow function.
- Return type
None- Returns
None
- dispatch(*args, **kwargs)[source]#
DEPRECATED: Function to dispatch workflows.
- Parameters
*args – Positional arguments for the workflow
**kwargs – Keyword arguments for the workflow
- Return type
str- Returns
Dispatch id assigned to job
- dispatch_sync(*args, **kwargs)[source]#
DEPRECATED: Function to dispatch workflows synchronously by waiting for the result too.
- Parameters
*args – Positional arguments for the workflow
**kwargs – Keyword arguments for the workflow
- Return type
- Returns
Result of workflow execution
- draw(*args, **kwargs)[source]#
Generate lattice graph and display in UI taking into account passed in arguments.
- Parameters
*args – Positional arguments to be passed to build the graph.
**kwargs – Keyword arguments to be passed to build the graph.
- Return type
None- Returns
None
Lepton#
Decorator to use languages other than Python, including scripting languages.
More robust definition of languages other than Python.
Dependencies#
Generic dependencies for an electron
- class covalent._workflow.deps.Deps(apply_fn=None, apply_args=[], apply_kwargs={}, *, retval_keyword='')[source]#
Generic dependency class used in specifying any kind of dependency for an electron.
- apply_fn#
function to be executed in the backend environment
- apply_args#
list of arguments to be applied in the backend environment
- apply_kwargs#
dictionary of keyword arguments to be applied in the backend environment
Methods:
apply()Encapsulates the exact function and args/kwargs to be executed in the backend environment.
- apply()[source]#
Encapsulates the exact function and args/kwargs to be executed in the backend environment.
- Parameters
None –
- Return type
Tuple[TransportableObject,TransportableObject,TransportableObject,str]- Returns
A tuple of transportable objects containing the function and optional args/kwargs
Add a Bash dependency to an electron
Add a callable dependency to an electron
Add a Pip dependency to an electron
Bash Dependencies#
Shell commands to run before an electron execution.
- class covalent._workflow.depsbash.DepsBash(commands=[])[source]#
Shell commands to run before an electron
Deps class to encapsulate Bash dependencies for an electron.
The specified commands will be executed as subprocesses in the same environment as the electron.
- commands#
A list of bash commands to execute before the electron runs.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Add a Bash dependency to an electron
Call Dependencies#
Functions, shell commands, PyPI packages, and other dependencies to be called in an electron’s execution environment.
- class covalent._workflow.depscall.DepsCall(func=None, args=None, kwargs=None, *, retval_keyword='', override_reserved_retval_keys=False)[source]#
Functions, shell commands, PyPI packages, and other types of dependencies to be called in an electron’s execution environment
Deps class to encapsulate python functions to be called in the same execution environment as the electron.
- func#
A callable
- args#
args list
- kwargs#
kwargs dict
- retval_keyword#
An optional string referencing the return value of func.
If retval_keyword is specified, the return value of func will be passed during workflow execution as an argument to the electron corresponding to the parameter of the same name.
Notes
Electron parameters to be injected during execution must have default parameter values.
It is the user’s responsibility to ensure that retval_keyword is actually a parameter of the electron. Unexpected behavior may occur otherwise.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Add a callable dependency to an electron
Pip Dependencies#
PyPI packages to be installed before executing an electron
- class covalent._workflow.depspip.DepsPip(packages=[], reqs_path='')[source]#
PyPI packages to be installed before executing an electron
A specification of Pip packages to be installed
- packages#
A list of PyPI packages to install
- reqs_path#
Path to requirements.txt (overrides packages)
These packages are installed in an electron’s execution environment just before the electron is run.
Methods:
from_dict(object_dict)Rehydrate a dictionary representation
to_dict()Return a JSON-serializable dictionary representation of self
Add a Pip dependency to an electron
File Transfer#
File Transfer from (source) and to (destination) local or remote files prior/post electron execution. Instances are are provided to files keyword argument in an electron decorator.
- class covalent._file_transfer.file.File(filepath=None, is_remote=False, is_dir=False, include_folder=False)[source]#
File class to store components of provided URI including scheme (s3://, file://, ect.) determine if the file is remote, and acts a facade to facilitate filesystem operations.
- filepath#
File path corresponding to the file.
- is_remote#
Flag determining if file is remote (override). Default is resolved automatically from file scheme.
- is_dir#
Flag determining if file is a directory (override). Default is determined if file uri contains trailing slash.
- include_folder#
Flag that determines if the folder should be included in the file transfer, if False only contents of folder are transfered.
- class covalent._file_transfer.folder.Folder(filepath=None, is_remote=False, is_dir=True, include_folder=False)[source]#
Folder class to store components of provided URI including scheme (s3://, file://, ect.), determine if the file is remote, and act as facade to facilitate filesystem operations. Folder is a child of the File class which sets is_dir flag to True.
- include_folder#
Flag that determines if the folder should be included in the file transfer, if False only contents of folder are transfered.
- class covalent._file_transfer.file_transfer.FileTransfer(from_file=None, to_file=None, order=<Order.BEFORE: 'before'>, strategy=None)[source]#
FileTransfer object class that takes two File objects or filepaths (from, to) and a File Transfer Strategy to perform remote or local file transfer operations.
- from_file#
Filepath or File object corresponding to the source file.
- to_file#
Filepath or File object corresponding to the destination file.
- order#
Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution.
- strategy#
Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.
- covalent._file_transfer.file_transfer.TransferFromRemote(from_filepath, to_filepath=None, strategy=None, order=<Order.BEFORE: 'before'>)[source]#
Factory for creating a FileTransfer instance where from_filepath is implicitly created as a remote File Object, and the order (Order.BEFORE) is set so that this file transfer will occur prior to electron execution.
- Parameters
from_filepath (
str) – File path corresponding to remote file (source).to_filepath (
Optional[str]) – File path corresponding to local file (destination)strategy (
Optional[FileTransferStrategy]) – Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.order (
Order) – Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution - default is BEFORE
- Return type
- Returns
FileTransfer instance with implicit Order.BEFORE enum set and from (source) file marked as remote
- covalent._file_transfer.file_transfer.TransferToRemote(to_filepath, from_filepath=None, strategy=None, order=<Order.AFTER: 'after'>)[source]#
Factory for creating a FileTransfer instance where to_filepath is implicitly created as a remote File Object, and the order (Order.AFTER) is set so that this file transfer will occur post electron execution.
- Parameters
to_filepath (
str) – File path corresponding to remote file (destination)from_filepath (
Optional[str]) – File path corresponding to local file (source).strategy (
Optional[FileTransferStrategy]) – Optional File Transfer Strategy to perform file operations - default will be resolved from provided file schemes.order (
Order) – Order (enum) to execute the file transfer before (Order.BEFORE) or after (Order.AFTER) electron execution - default is AFTER
- Return type
- Returns
FileTransfer instance with implicit Order.AFTER enum set and to (destination) file marked as remote
Transfer files to and from a remote host
File Transfer Strategies#
A set of classes that support various protocols. All FileTransferStrategy classes share an interface to perform copy, download, and upload operations on two File objects (a source and a destination).
Transfer files to and from a remote host
Synchronous Base Executor Class#
- class covalent.executor.base.BaseExecutor(*args, **kwargs)[source]#
Base executor class to be used for defining any executor plugin. Subclassing this class will allow you to define your own executor plugin which can be used in covalent.
- log_stdout#
The path to the file to be used for redirecting stdout.
- log_stderr#
The path to the file to be used for redirecting stderr.
- cache_dir#
The location used for cached files in the executor.
- time_limit#
time limit for the task
- retries#
Number of times to retry execution upon failure
Methods:
cancel(task_metadata, job_handle)Method to cancel the job identified uniquely by the job_handle (base class)
execute(function, args, kwargs, dispatch_id, …)Execute the function with the given arguments.
from_dict(object_dict)Rehydrate a dictionary representation
Check if the task was requested to be cancelled by the user
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
Query the database for the task’s Python and Covalent version
run(function, args, kwargs, task_metadata)Abstract method to run a function in the executor.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job_id/handle returned by the backend executing the task
set_job_status(status)Sets the job state
setup(task_metadata)Placeholder to run any executor specific tasks
teardown(task_metadata)Placeholder to run any executor specific cleanup/teardown actions
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- cancel(task_metadata, job_handle)[source]#
Method to cancel the job identified uniquely by the job_handle (base class)
- Arg(s)
task_metadata: Metadata of the task to be cancelled job_handle: Unique ID of the job assigned by the backend
- Return(s)
False by default
- Return type
bool
- execute(function, args, kwargs, dispatch_id, results_dir, node_id=- 1)[source]#
Execute the function with the given arguments.
This calls the executor-specific run() method.
- Parameters
function (
Callable) – The input python function which will be executed and whose result is ultimately returned by this function.args (
List) – List of positional arguments to be used by the function.kwargs (
Dict) – Dictionary of keyword arguments to be used by the function.dispatch_id (
str) – The unique identifier of the external lattice process which is calling this function.results_dir (
str) – The location of the results directory.node_id (
int) – ID of the node in the transport graph which is using this executor.
- Returns
The result of the function execution.
- Return type
output
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- get_cancel_requested()[source]#
Check if the task was requested to be cancelled by the user
- Arg(s)
None
- Return(s)
True/False whether task cancellation was requested
- Return type
bool
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- get_version_info()[source]#
Query the database for the task’s Python and Covalent version
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- abstract run(function, args, kwargs, task_metadata)[source]#
Abstract method to run a function in the executor.
- Parameters
function (
Callable) – The function to run in the executorargs (
List) – List of positional arguments to be used by the functionkwargs (
Dict) – Dictionary of keyword arguments to be used by the function.task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id
- Returns
The result of the function execution
- Return type
output
- async send(task_specs, resources, task_group_metadata)[source]#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[Dict]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – a dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- set_job_handle(handle)[source]#
Save the job_id/handle returned by the backend executing the task
- Arg(s)
handle: Any JSONable type to identifying the task being executed by the backend
- Return(s)
Response from saving the job handle to database
- Return type
Any
- async set_job_status(status)[source]#
Sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- teardown(task_metadata)[source]#
Placeholder to run any executor specific cleanup/teardown actions
- Return type
Any
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)[source]#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
- Return type
None
Asynchronous Base Executor Class#
- class covalent.executor.base.AsyncBaseExecutor(*args, **kwargs)[source]#
Async base executor class to be used for defining any executor plugin. Subclassing this class will allow you to define your own executor plugin which can be used in covalent.
This is analogous to BaseExecutor except the run() method, together with the optional setup() and teardown() methods, are coroutines.
- log_stdout#
The path to the file to be used for redirecting stdout.
- log_stderr#
The path to the file to be used for redirecting stderr.
- cache_dir#
The location used for cached files in the executor.
- time_limit#
time limit for the task
- retries#
Number of times to retry execution upon failure
Methods:
cancel(task_metadata, job_handle)Method to cancel the job identified uniquely by the job_handle (base class)
from_dict(object_dict)Rehydrate a dictionary representation
Get if the task was requested to be canceled
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
Query the database for dispatch version metadata.
poll(task_group_metadata, data)Block until the job has reached a terminal state.
receive(task_group_metadata, data)Return a list of task updates.
run(function, args, kwargs, task_metadata)Abstract method to run a function in the executor in async-aware manner.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job handle to database
set_job_status(status)Validates and sets the job state
setup(task_metadata)Executor specific setup method
teardown(task_metadata)Executor specific teardown method
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- async cancel(task_metadata, job_handle)[source]#
Method to cancel the job identified uniquely by the job_handle (base class)
- Arg(s)
task_metadata: Metadata of the task to be cancelled job_handle: Unique ID of the job assigned by the backend
- Return(s)
False by default
- Return type
bool
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- async get_cancel_requested()[source]#
Get if the task was requested to be canceled
- Arg(s)
None
- Return(s)
Whether the task has been requested to be cancelled
- Return type
Any
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- async get_version_info()[source]#
Query the database for dispatch version metadata.
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- async poll(task_group_metadata, data)[source]#
Block until the job has reached a terminal state.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of send().
The return value of poll() will be passed directly into receive().
Raise NotImplementedError to indicate that the compute backend will notify the Covalent server asynchronously of job completion.
- Return type
Any
- async receive(task_group_metadata, data)[source]#
Return a list of task updates.
Each task must have reached a terminal state by the time this is invoked.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of poll() or the request body of /jobs/update.
- Return type
List[TaskUpdate]- Returns
Returns a list of task results, each a TaskUpdate dataclass of the form
- {
“dispatch_id”: dispatch_id, “node_id”: node_id, “status”: status, “assets”: {
- ”output”: {
“remote_uri”: output_uri,
}, “stdout”: {
”remote_uri”: stdout_uri,
}, “stderr”: {
”remote_uri”: stderr_uri,
},
},
}
corresponding to the node ids (task_ids) specified in the task_group_metadata. This might be a subset of the node ids in the originally submitted task group as jobs may notify Covalent asynchronously of completed tasks before the entire task group finishes running.
- abstract async run(function, args, kwargs, task_metadata)[source]#
Abstract method to run a function in the executor in async-aware manner.
- Parameters
function (
Callable) – The function to run in the executorargs (
List) – List of positional arguments to be used by the functionkwargs (
Dict) – Dictionary of keyword arguments to be used by the function.task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id
- Returns
The result of the function execution
- Return type
output
- async send(task_specs, resources, task_group_metadata)[source]#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[TaskSpec]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- Return type
Any
- async set_job_handle(handle)[source]#
Save the job handle to database
- Arg(s)
handle: JSONable type identifying the job being executed by the backend
- Return(s)
Response from the listener that handles inserting the job handle to database
- Return type
Any
- async set_job_status(status)[source]#
Validates and sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- async write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)[source]#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
This uses aiofiles to avoid blocking the event loop.
- Return type
None
Dask Executor#
Executing tasks (electrons) in a Dask cluster. This is the default executor when covalent is started without the --no-cluster flag.
from dask.distributed import LocalCluster
cluster = LocalCluster()
print(cluster.scheduler_address)
The address will look like tcp://127.0.0.1:55564 when running locally. Note that the Dask cluster does not persist when the process terminates.
This cluster can be used with Covalent by providing the scheduler address:
import covalent as ct
dask_executor = ct.executor.DaskExecutor(
scheduler_address="tcp://127.0.0.1:55564"
)
@ct.electron(executor=dask_executor)
def my_custom_task(x, y):
return x + y
...
Local Executor#
Executing tasks (electrons) directly on the local machine
AWS Plugins#
Covalent is a python based workflow orchestration tool used to execute HPC and quantum tasks in heterogenous environments.
By installing Covalent AWS Plugins users can leverage a broad plugin ecosystem to execute tasks using AWS resources best fit for each task.
Covalent AWS Plugins installs a set of executor plugins that allow tasks to be run in an EC2 instance, AWS Lambda, AWS ECS Cluster, AWS Batch Compute Environment, and as an AWS Braket Job for tasks requiring Quantum devices.
If you’re new to Covalent see the Getting Started Guide.
To use the AWS plugin ecosystem with Covalent, simply install it with pip:
pip install "covalent-aws-plugins[all]"
This will ensure that all the AWS executor plugins listed below are installed.
Note
Users will require Terraform to be installed in order to use the EC2 plugin.
While each plugin can be seperately installed installing the above pip package installs all of the below plugins.
Plugin Name |
Use Case |
|
|---|---|---|

|
AWS Batch Executor |
Useful for heavy compute workloads (high CPU/memory). Tasks are queued to execute in the user defined Batch compute environment. |

|
AWS EC2 Executor |
General purpose compute workloads where users can select compute resources. An EC2 instance is auto-provisioned using terraform with selected compute settings to execute tasks. |

|
AWS Braket Executor |
Suitable for Quantum/Classical hybrid workflows. Tasks are executed using a combination of classical and quantum devices. |

|
AWS ECS Executor |
Useful for moderate to heavy workloads (low memory requirements). Tasks are executed in an AWS ECS cluster as containers. |

|
AWS Lambda Executor |
Suitable for short lived tasks that can be parallalized (low memory requirements). Tasks are executed in serverless AWS Lambda functions. |
Firstly, import covalent
import covalent as ct
Secondly, define your executor
executor = ct.executor.AWSBatchExecutor(
s3_bucket_name = "covalent-batch-qa-job-resources",
batch_job_definition_name = "covalent-batch-qa-job-definition",
batch_queue = "covalent-batch-qa-queue",
batch_execution_role_name = "ecsTaskExecutionRole",
batch_job_role_name = "covalent-batch-qa-job-role",
batch_job_log_group_name = "covalent-batch-qa-log-group",
vcpu = 2, # Number of vCPUs to allocate
memory = 3.75, # Memory in GB to allocate
time_limit = 300, # Time limit of job in seconds
)
executor = ct.executor.EC2Executor(
instance_type="t2.micro",
volume_size=8, #GiB
ssh_key_file="~/.ssh/ec2_key"
)
executor = ct.executor.BraketExecutor(
s3_bucket_name="braket_s3_bucket",
ecr_repo_name="braket_ecr_repo",
braket_job_execution_role_name="covalent-braket-iam-role",
quantum_device="arn:aws:braket:::device/quantum-simulator/amazon/sv1",
classical_device="ml.m5.large",
storage=30,
)
executor = ct.executor.ECSExecutor(
s3_bucket_name="covalent-fargate-task-resources",
ecr_repo_name="covalent-fargate-task-images",
ecs_cluster_name="covalent-fargate-cluster",
ecs_task_family_name="covalent-fargate-tasks",
ecs_task_execution_role_name="ecsTaskExecutionRole",
ecs_task_role_name="CovalentFargateTaskRole",
ecs_task_subnet_id="subnet-000000e0",
ecs_task_security_group_id="sg-0000000a",
ecs_task_log_group_name="covalent-fargate-task-logs",
vcpu=1,
memory=2
)
executor = ct.executor.AWSLambdaExecutor(
lambda_role_name="CovalentLambdaExecutionRole",
s3_bucket_name="covalent-lambda-job-resources",
timeout=60,
memory_size=512
)
Lastly, define a workflow to execute a particular task using one of the above executors
@ct.electron(
executor=executor
)
def compute_pi(n):
# Leibniz formula for π
return 4 * sum(1.0/(2*i + 1)*(-1)**i for i in range(n))
@ct.lattice
def workflow(n):
return compute_pi(n)
dispatch_id = ct.dispatch(workflow)(100000000)
result = ct.get_result(dispatch_id=dispatch_id, wait=True)
print(result.result)
Which should output
3.141592643589326
AWS Batch Executor#
Covalent is a Pythonic workflow tool used to execute tasks on advanced computing hardware.
This executor plugin interfaces Covalent with AWS Batch which allows tasks in a covalent workflow to be executed as AWS batch jobs.
Furthermore, this plugin is well suited for compute/memory intensive tasks such as training machine learning models, hyperparameter optimization, deep learning etc. With this executor, the compute backend is the Amazon EC2 service, with instances optimized for compute and memory intensive operations.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-awsbatch-plugin
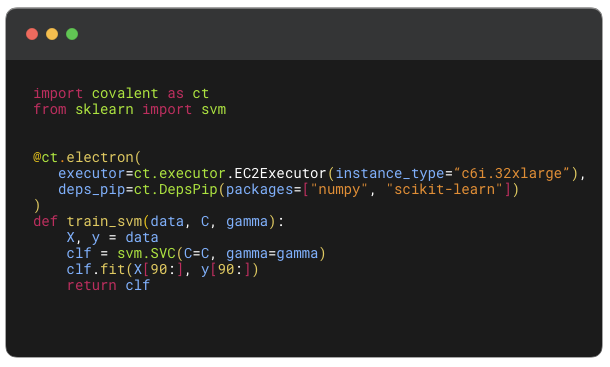
This is an example of how a workflow can be adapted to utilize the AWS Batch Executor. Here we train a simple Support Vector Machine (SVM) model and use an existing AWS Batch Compute environment to run the train_svm electron as a batch job. We also note we require DepsPip to install the dependencies when creating the batch job.
from numpy.random import permutation
from sklearn import svm, datasets
import covalent as ct
deps_pip = ct.DepsPip(
packages=["numpy==1.23.2", "scikit-learn==1.1.2"]
)
executor = ct.executor.AWSBatchExecutor(
s3_bucket_name = "covalent-batch-qa-job-resources",
batch_job_definition_name = "covalent-batch-qa-job-definition",
batch_queue = "covalent-batch-qa-queue",
batch_execution_role_name = "ecsTaskExecutionRole",
batch_job_role_name = "covalent-batch-qa-job-role",
batch_job_log_group_name = "covalent-batch-qa-log-group",
vcpu = 2, # Number of vCPUs to allocate
memory = 3.75, # Memory in GB to allocate
time_limit = 300, # Time limit of job in seconds
)
# Use executor plugin to train our SVM model.
@ct.electron(
executor=executor,
deps_pip=deps_pip
)
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
@ct.electron
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
@ct.electron
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],
y_test[:90]
)
@ct.lattice
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(
data=data,
clf=clf
)
return score
# Dispatch the workflow
dispatch_id = ct.dispatch(run_experiment)(
C=1.0,
gamma=0.7
)
# Wait for our result and get result value
result = ct.get_result(dispatch_id=dispatch_id, wait=True).result
print(result)
During the execution of the workflow one can navigate to the UI to see the status of the workflow, once completed however the above script should also output a value with the score of our model.
0.8666666666666667
Config Key |
Is Required |
Default |
Description |
|---|---|---|---|
profile |
No |
default |
Named AWS profile used for authentication |
region |
Yes |
us-east-1 |
AWS Region to use to for client calls |
credentials |
No |
~/.aws/credentials |
The path to the AWS credentials file |
batch_queue |
Yes |
covalent-batch-queue |
Name of the Batch queue used for job management. |
s3_bucket_name |
Yes |
covalent-batch-job-resources |
Name of an S3 bucket where covalent artifacts are stored. |
batch_job_definition_name |
Yes |
covalent-batch-jobs |
Name of the Batch job definition for a user, project, or experiment. |
batch_execution_role_name |
No |
ecsTaskExecutionRole |
Name of the IAM role used by the Batch ECS agent (the above role should already exist in AWS). |
batch_job_role_name |
Yes |
CovalentBatchJobRole |
Name of the IAM role used within the container. |
batch_job_log_group_name |
Yes |
covalent-batch-job-logs |
Name of the CloudWatch log group where container logs are stored. |
vcpu |
No |
2 |
Number of vCPUs available to a task. |
memory |
No |
3.75 |
Memory (in GB) available to a task. |
num_gpus |
No |
0 |
Number of GPUs availabel to a task. |
retry_attempts |
No |
3 |
Number of times a job is retried if it fails. |
time_limit |
No |
300 |
Time limit (in seconds) after which jobs are killed. |
poll_freq |
No |
10 |
Frequency (in seconds) with which to poll a submitted task. |
cache_dir |
No |
/tmp/covalent |
Cache directory used by this executor for temporary files. |
This plugin can be configured in one of two ways:
Configuration options can be passed in as constructor keys to the executor class
ct.executor.AWSBatchExecutorBy modifying the covalent configuration file under the section
[executors.awsbatch]
The following shows an example of how a user might modify their covalent configuration file to support this plugin:
[executors.awsbatch]
s3_bucket_name = "covalent-batch-job-resources"
batch_queue = "covalent-batch-queue"
batch_job_definition_name = "covalent-batch-jobs"
batch_execution_role_name = "ecsTaskExecutionRole"
batch_job_role_name = "CovalentBatchJobRole"
batch_job_log_group_name = "covalent-batch-job-logs"
...
In order to run your workflows with covalent there are a few notable AWS resources that need to be provisioned first.
Resource |
Is Required |
Config Key |
Description |
|---|---|---|---|
AWS S3 Bucket |
Yes |
|
S3 bucket must be created for covalent to store essential files that are needed during execution. |
VPC & Subnet |
Yes |
N/A |
A VPC must be associated with the AWS Batch Compute Environment along with a public or private subnet (there needs to be additional resources created for private subnets) |
AWS Batch Compute Environment |
Yes |
N/A |
An AWS Batch compute environment (EC2) that will provision EC2 instances as needed when jobs are submitted to the associated job queue. |
AWS Batch Queue |
Yes |
|
An AWS Batch Job Queue that will queue tasks for execution in it’s associated compute environment. |
AWS Batch Job Definition |
Yes |
|
An AWS Batch job definition that will be replaced by a new batch job definition when the workflow is executed. |
AWS IAM Role (Job Role) |
Yes |
|
The IAM role used within the container. |
AWS IAM Role (Execution Role) |
No |
|
The IAM role used by the Batch ECS agent (default role ecsTaskExecutionRole should already exist). |
Log Group |
Yes |
|
An AWS CloudWatch log group where task logs are stored. |
To create an AWS S3 Bucket refer to the following AWS documentation.
To create a VPC & Subnet refer to the following AWS documentation.
To create an AWS Batch Queue refer to the following AWS documentation it must be a compute environment configured in EC2 mode.
To create an AWS Batch Job Definition refer to the following AWS documentation the configuration for this can be trivial as covalent will update the Job Definition prior to execution.
To create an AWS IAM Role for batch jobs (Job Role) one can provision a policy with the following permissions (below) then create a new role and attach with the created policy. Refer to the following AWS documentation for an example of creating a policy & role in IAM.
AWS Batch IAM Job Policy
{
"Version": "2012-10-17",
"Statement": [
{
"Sid": "BatchJobMgmt",
"Effect": "Allow",
"Action": [
"batch:TerminateJob",
"batch:DescribeJobs",
"batch:SubmitJob",
"batch:RegisterJobDefinition"
],
"Resource": "*"
},
{
"Sid": "ECRAuth",
"Effect": "Allow",
"Action": [
"ecr:GetAuthorizationToken"
],
"Resource": "*"
},
{
"Sid": "ECRUpload",
"Effect": "Allow",
"Action": [
"ecr:GetDownloadUrlForLayer",
"ecr:BatchGetImage",
"ecr:BatchCheckLayerAvailability",
"ecr:InitiateLayerUpload",
"ecr:UploadLayerPart",
"ecr:CompleteLayerUpload",
"ecr:PutImage"
],
"Resource": [
"arn:aws:ecr:<region>:<account>:repository/<ecr_repo_name>"
]
},
{
"Sid": "IAMRoles",
"Effect": "Allow",
"Action": [
"iam:GetRole",
"iam:PassRole"
],
"Resource": [
"arn:aws:iam::<account>:role/CovalentBatchJobRole",
"arn:aws:iam::<account>:role/ecsTaskExecutionRole"
]
},
{
"Sid": "ObjectStore",
"Effect": "Allow",
"Action": [
"s3:ListBucket",
"s3:PutObject",
"s3:GetObject"
],
"Resource": [
"arn:aws:s3:::<s3_resource_bucket>/*",
"arn:aws:s3:::<s3_resource_bucket>"
]
},
{
"Sid": "LogRead",
"Effect": "Allow",
"Action": [
"logs:GetLogEvents"
],
"Resource": [
"arn:aws:logs:<region>:<account>:log-group:<cloudwatch_log_group_name>:log-stream:*"
]
}
]
}
- class covalent_awsbatch_plugin.awsbatch.AWSBatchExecutor(credentials=None, profile=None, region=None, s3_bucket_name=None, batch_queue=None, batch_execution_role_name=None, batch_job_role_name=None, batch_job_log_group_name=None, vcpu=None, memory=None, num_gpus=None, retry_attempts=None, time_limit=None, poll_freq=None, cache_dir=None, container_image_uri=None)[source]#
AWS Batch executor plugin class.
- Parameters
credentials (
Optional[str]) – Full path to AWS credentials file.profile (
Optional[str]) – Name of an AWS profile whose credentials are used.s3_bucket_name (
Optional[str]) – Name of an S3 bucket where objects are stored.batch_queue (
Optional[str]) – Name of the Batch queue used for job management.batch_execution_role_name (
Optional[str]) – Name of the IAM role used by the Batch ECS agent.batch_job_role_name (
Optional[str]) – Name of the IAM role used within the container.batch_job_log_group_name (
Optional[str]) – Name of the CloudWatch log group where container logs are stored.vcpu (
Optional[int]) – Number of vCPUs available to a task.memory (
Optional[float]) – Memory (in GB) available to a task.num_gpus (
Optional[int]) – Number of GPUs available to a task.retry_attempts (
Optional[int]) – Number of times a job is retried if it fails.time_limit (
Optional[int]) – Time limit (in seconds) after which jobs are killed.poll_freq (
Optional[int]) – Frequency with which to poll a submitted task.cache_dir (
Optional[str]) – Cache directory used by this executor for temporary files.container_image_uri (
Optional[str]) – URI of the docker container image used by the executor.
Methods:
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
cancel(task_metadata, job_handle)Cancel the batch job.
from_dict(object_dict)Rehydrate a dictionary representation
Get if the task was requested to be canceled
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
get_status(job_id)Query the status of a previously submitted Batch job.
Query the database for dispatch version metadata.
poll(task_group_metadata, data)Block until the job has reached a terminal state.
query_result(task_metadata)Query and retrieve a completed job’s result.
receive(task_group_metadata, data)Return a list of task updates.
run(function, args, kwargs, task_metadata)Abstract method to run a function in the executor in async-aware manner.
run_async_subprocess(cmd)Invokes an async subprocess to run a command.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job handle to database
set_job_status(status)Validates and sets the job state
setup(task_metadata)Executor specific setup method
submit_task(task_metadata, identity)Invokes the task on the remote backend.
teardown(task_metadata)Executor specific teardown method
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- boto_session_options()#
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
- Return type
Dict[str,str]
- async cancel(task_metadata, job_handle)[source]#
Cancel the batch job.
- Parameters
task_metadata (
Dict) – Dictionary with the task’s dispatch_id and node id.job_handle (
str) – Unique job handle assigned to the task by AWS Batch.
- Return type
bool- Returns
If the job was cancelled or not
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- async get_cancel_requested()#
Get if the task was requested to be canceled
- Arg(s)
None
- Return(s)
Whether the task has been requested to be cancelled
- Return type
Any
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- async get_status(job_id)[source]#
Query the status of a previously submitted Batch job.
- Parameters
batch – Batch client object.
job_id (
str) – Identifier used to identify a Batch job.
- Returns
String describing the task status. exit_code: Exit code, if the task has completed, else -1.
- Return type
status
- async get_version_info()#
Query the database for dispatch version metadata.
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- async poll(task_group_metadata, data)#
Block until the job has reached a terminal state.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of send().
The return value of poll() will be passed directly into receive().
Raise NotImplementedError to indicate that the compute backend will notify the Covalent server asynchronously of job completion.
- Return type
Any
- async query_result(task_metadata)[source]#
Query and retrieve a completed job’s result.
- Parameters
task_metadata (
Dict) – Dictionary containing the task dispatch_id and node_id- Return type
Tuple[Any,str,str]- Returns
result, stdout, stderr
- async receive(task_group_metadata, data)#
Return a list of task updates.
Each task must have reached a terminal state by the time this is invoked.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of poll() or the request body of /jobs/update.
- Return type
List[TaskUpdate]- Returns
Returns a list of task results, each a TaskUpdate dataclass of the form
- {
“dispatch_id”: dispatch_id, “node_id”: node_id, “status”: status, “assets”: {
- ”output”: {
“remote_uri”: output_uri,
}, “stdout”: {
”remote_uri”: stdout_uri,
}, “stderr”: {
”remote_uri”: stderr_uri,
},
},
}
corresponding to the node ids (task_ids) specified in the task_group_metadata. This might be a subset of the node ids in the originally submitted task group as jobs may notify Covalent asynchronously of completed tasks before the entire task group finishes running.
- async run(function, args, kwargs, task_metadata)[source]#
Abstract method to run a function in the executor in async-aware manner.
- Parameters
function (
Callable) – The function to run in the executorargs (
List) – List of positional arguments to be used by the functionkwargs (
Dict) – Dictionary of keyword arguments to be used by the function.task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id
- Returns
The result of the function execution
- Return type
output
- async static run_async_subprocess(cmd)#
Invokes an async subprocess to run a command.
- Return type
Tuple
- async send(task_specs, resources, task_group_metadata)#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[TaskSpec]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- Return type
Any
- async set_job_handle(handle)#
Save the job handle to database
- Arg(s)
handle: JSONable type identifying the job being executed by the backend
- Return(s)
Response from the listener that handles inserting the job handle to database
- Return type
Any
- async set_job_status(status)#
Validates and sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- async setup(task_metadata)#
Executor specific setup method
- async submit_task(task_metadata, identity)[source]#
Invokes the task on the remote backend.
- Parameters
task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id.identity (
Dict) – Dictionary from _validate_credentials call { “Account”: “AWS Account ID”, …}
- Returns
Task UUID defined on the remote backend.
- Return type
task_uuid
- async teardown(task_metadata)#
Executor specific teardown method
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- validate_status(status)#
Overridable filter
- Return type
bool
- async write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
This uses aiofiles to avoid blocking the event loop.
- Return type
None
AWS Braket Executor#
Covalent is a Pythonic workflow tool used to execute tasks on advanced computing hardware.
This plugin allows executing quantum circuits and quantum-classical hybrid jobs in Amazon Braket when you use Covalent.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-braket-plugin
The following toy example executes a simple quantum circuit on one qubit that prepares a uniform superposition of the standard basis states and then measures the state. We use the Pennylane framework.
import covalent as ct
from covalent_braket_plugin.braket import BraketExecutor
import os
# AWS resources to pass to the executor
credentials = "~/.aws/credentials"
profile = "default"
region = "us-east-1"
s3_bucket_name = "braket_s3_bucket"
ecr_repo_name = "braket_ecr_repo"
iam_role_name = "covalent-braket-iam-role"
# Instantiate the executor
ex = BraketExecutor(
profile=profile,
credentials=credentials_file,
s3_bucket_name=s3_bucket_name,
ecr_image_uri=ecr_image_uri,
braket_job_execution_role_name=iam_role_name,
quantum_device="arn:aws:braket:::device/quantum-simulator/amazon/sv1",
classical_device="ml.m5.large",
storage=30,
time_limit=300,
)
# Execute the following circuit:
# |0> - H - Measure
@ct.electron(executor=ex)
def simple_quantum_task(num_qubits: int):
import pennylane as qml
# These are passed to the Hybrid Jobs container at runtime
device_arn = os.environ["AMZN_BRAKET_DEVICE_ARN"]
s3_bucket = os.environ["AMZN_BRAKET_OUT_S3_BUCKET"]
s3_task_dir = os.environ["AMZN_BRAKET_TASK_RESULTS_S3_URI"].split(s3_bucket)[1]
device = qml.device(
"braket.aws.qubit",
device_arn=device_arn,
s3_destination_folder=(s3_bucket, s3_task_dir),
wires=num_qubits,
)
@qml.qnode(device=device)
def simple_circuit():
qml.Hadamard(wires=[0])
return qml.expval(qml.PauliZ(wires=[0]))
res = simple_circuit().numpy()
return res
@ct.lattice
def simple_quantum_workflow(num_qubits: int):
return simple_quantum_task(num_qubits=num_qubits)
dispatch_id = ct.dispatch(simple_quantum_workflow)(1)
result_object = ct.get_result(dispatch_id, wait=True)
# We expect 0 as the result
print("Result:", result_object.result)
During the execution of the workflow one can navigate to the UI to see the status of the workflow, once completed however the above script should also output a value with the output of the quantum measurement.
>>> Result: 0
Config Key |
Is Required |
Default |
Description |
|---|---|---|---|
credentials |
No |
“~/.aws/credentials” |
The path to the AWS credentials file |
braket_job_execution_role_name |
Yes |
“CovalentBraketJobsExecutionRole” |
The name of the IAM role that Braket will assume during task execution. |
profile |
No |
“default” |
Named AWS profile used for authentication |
region |
Yes |
:code`AWS_DEFAULT_REGION` environment variable |
AWS Region to use to for client calls to AWS |
s3_bucket_name |
Yes |
amazon-braket-covalent-job-resources |
The S3 bucket where Covalent will store input and output files for the task. |
ecr_image_uri |
Yes |
An ECR repository for storing container images to be run by Braket. |
|
quantum_device |
No |
“arn:aws:braket:::device/quantum-simulator/amazon/sv1” |
The ARN of the quantum device to use |
classical_device |
No |
“ml.m5.large” |
Instance type for the classical device to use |
storage |
No |
30 |
Storage size in GB for the classical device |
time_limit |
No |
300 |
Max running time in seconds for the Braket job |
poll_freq |
No |
30 |
How often (in seconds) to poll Braket for the job status |
cache_dir |
No |
“/tmp/covalent” |
Location for storing temporary files generated by the Covalent server |
This plugin can be configured in one of two ways:
Configuration options can be passed in as constructor keys to the executor class
ct.executor.BraketExecutorBy modifying the covalent configuration file under the section
[executors.braket]
The following shows an example of how a user might modify their covalent configuration file to support this plugin:
[executors.braket]
quantum_device = "arn:aws:braket:::device/qpu/ionq/ionQdevice"
time_limit = 3600
The Braket executor requires some resources to be provisioned on AWS. Precisely, users will need an S3 bucket, an ECR repo, and an IAM role with the appropriate permissions to be passed to Braket.
Resource |
Is Required |
Config Key |
Description |
|---|---|---|---|
IAM role |
Yes |
|
An IAM role granting permissions to Braket, S3, ECR, and a few other resources. |
ECR repository |
Yes |
|
An ECR repository for storing container images to be run by Braket. |
S3 bucket |
Yes |
|
An S3 bucket for storing task-specific data, such as Braket outputs or function inputs. |
One can either follow the below instructions to manually create the resources or use the provided terraform script to auto-provision the resources needed.
The AWS documentation on S3 details how to configure an S3 bucket.
The permissions required for the the IAM role are documented in the article “managing access to Amazon Braket”. The following policy is attached to the default role “CovalentBraketJobsExecutionRole”:
In order to use the Braket executor plugin one must create a private ECR registry with a container image that will be used to execute the Braket jobs using covalent. One can either create an ECR repository manually or use the terraform script provided below. We host the image in our public repository at
public.ecr.aws/covalent/covalent-braket-executor:stable
Note
The container image can be uploaded to a private ECR as follows
docker pull public.ecr.aws/covalent/covalent-braket-executor:stable
Once the image has been obtained, user’s can tag it with their registry information and upload to ECR as follows
aws ecr get-login-password --region <region> | docker login --username AWS --password-stdin <aws_account_id>.dkr.ecr.<region>.amazonaws.com
docker tag public.ecr.aws/covalent/covalent-braket-executor:stable <aws_account_id>.dkr.ecr.<region>.amazonaws.com/<my-repository>:tag
docker push <aws_account_id>.dkr.ecr.<region>.amazonaws.com/<my-repository>:tag
Sample IAM policy for Braket’s execution role
- {
“Version”: “2012-10-17”, “Statement”: [
- {
“Sid”: “VisualEditor0”, “Effect”: “Allow”, “Action”: “cloudwatch:PutMetricData”, “Resource”: “*”, “Condition”: {
“StringEquals”: { “cloudwatch:namespace”: “/aws/braket” }
}
}, {
“Sid”: “VisualEditor1”, “Effect”: “Allow”, “Action”: [
“logs:CreateLogStream”, “logs:DescribeLogStreams”, “ecr:GetDownloadUrlForLayer”, “ecr:BatchGetImage”, “logs:StartQuery”, “logs:GetLogEvents”, “logs:CreateLogGroup”, “logs:PutLogEvents”, “ecr:BatchCheckLayerAvailability”
], “Resource”: [
“arn:aws:ecr::348041629502:repository/”, “arn:aws:logs:::log-group:/aws/braket*”
]
}, {
“Sid”: “VisualEditor2”, “Effect”: “Allow”, “Action”: “iam:PassRole”, “Resource”: “arn:aws:iam::348041629502:role/CovalentBraketJobsExecutionRole”, “Condition”: {
“StringLike”: { “iam:PassedToService”: “braket.amazonaws.com” }
}
}, {
“Sid”: “VisualEditor3”, “Effect”: “Allow”, “Action”: [
“braket:SearchDevices”, “s3:CreateBucket”, “ecr:BatchDeleteImage”, “ecr:BatchGetRepositoryScanningConfiguration”, “ecr:DeleteRepository”, “ecr:TagResource”, “ecr:BatchCheckLayerAvailability”, “ecr:GetLifecyclePolicy”, “braket:CreateJob”, “ecr:DescribeImageScanFindings”, “braket:GetJob”, “ecr:CreateRepository”, “ecr:PutImageScanningConfiguration”, “ecr:GetDownloadUrlForLayer”, “ecr:DescribePullThroughCacheRules”, “ecr:GetAuthorizationToken”, “ecr:DeleteLifecyclePolicy”, “braket:ListTagsForResource”, “ecr:PutImage”, “s3:PutObject”, “s3:GetObject”, “braket:GetDevice”, “ecr:UntagResource”, “ecr:BatchGetImage”, “ecr:DescribeImages”, “braket:CancelQuantumTask”, “ecr:StartLifecyclePolicyPreview”, “braket:CancelJob”, “ecr:InitiateLayerUpload”, “ecr:PutImageTagMutability”, “ecr:StartImageScan”, “ecr:DescribeImageReplicationStatus”, “ecr:ListTagsForResource”, “s3:ListBucket”, “ecr:UploadLayerPart”, “ecr:CreatePullThroughCacheRule”, “ecr:ListImages”, “ecr:GetRegistryScanningConfiguration”, “braket:TagResource”, “ecr:CompleteLayerUpload”, “ecr:DescribeRepositories”, “ecr:ReplicateImage”, “ecr:GetRegistryPolicy”, “ecr:PutLifecyclePolicy”, “s3:PutBucketPublicAccessBlock”, “ecr:GetLifecyclePolicyPreview”, “ecr:DescribeRegistry”, “braket:SearchJobs”, “braket:CreateQuantumTask”, “iam:ListRoles”, “ecr:PutRegistryScanningConfiguration”, “ecr:DeletePullThroughCacheRule”, “braket:UntagResource”, “ecr:BatchImportUpstreamImage”, “braket:GetQuantumTask”, “s3:PutBucketPolicy”, “braket:SearchQuantumTasks”, “ecr:GetRepositoryPolicy”, “ecr:PutReplicationConfiguration”
], “Resource”: “*”
}, {
“Sid”: “VisualEditor4”, “Effect”: “Allow”, “Action”: “logs:GetQueryResults”, “Resource”: “arn:aws:logs:::log-group:*”
}, {
“Sid”: “VisualEditor5”, “Effect”: “Allow”, “Action”: “logs:StopQuery”, “Resource”: “arn:aws:logs:::log-group:/aws/braket*”
}
]
}
Users can use the following Terraform snippet as a starting point to spin up the required resources
provider "aws" {}
data "aws_caller_identity" "current" {}
resource "aws_s3_bucket" "braket_bucket" {
bucket = "my-s3-bucket-name"
force_destroy = true
}
resource "aws_ecr_repository" "braket_ecr_repo" {
name = "amazon-braket-base-executor-repo"
image_tag_mutability = "MUTABLE"
force_delete = true
image_scanning_configuration {
scan_on_push = false
}
provisioner "local-exec" {
command = "docker pull public.ecr.aws/covalent/covalent-braket-executor:stable && aws ecr get-login-password --region <region> | docker login --username AWS --password-stdin ${data.aws_caller_identity.current.account_id}.dkr.ecr.${var.aws_region}.amazonaws.com && docker tag public.ecr.aws/covalent/covalent-braket-executor:stable ${aws_ecr_repository.braket_ecr_repo.repository_url}:stable && docker push ${aws_ecr_repository.braket_ecr_repo.repository_url}:stable"
}
}
resource "aws_iam_role" "braket_iam_role" {
name = "amazon-braket-execution-role"
assume_role_policy = jsonencode({
Version = "2012-10-17"
Statement = [
{
Action = "sts:AssumeRole"
Effect = "Allow"
Sid = ""
Principal = {
Service = "braket.amazonaws.com"
}
},
]
})
managed_policy_arns = ["arn:aws:iam::aws:policy/AmazonBraketFullAccess"]
}
- class covalent_braket_plugin.braket.BraketExecutor(ecr_image_uri=None, s3_bucket_name=None, braket_job_execution_role_name=None, classical_device=None, storage=None, time_limit=None, poll_freq=None, quantum_device=None, profile=None, credentials=None, cache_dir=None, region=None, **kwargs)[source]#
AWS Braket Hybrid Jobs executor plugin class.
Methods:
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
cancel()Abstract method that sends a cancellation request to the remote backend.
from_dict(object_dict)Rehydrate a dictionary representation
Get if the task was requested to be canceled
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
get_status(braket, job_arn)Query the status of a previously submitted Braket hybrid job.
Query the database for dispatch version metadata.
poll(task_group_metadata, data)Block until the job has reached a terminal state.
query_result(query_metadata)Abstract method that retrieves the pickled result from the remote cache.
receive(task_group_metadata, data)Return a list of task updates.
run(function, args, kwargs, task_metadata)Abstract method to run a function in the executor in async-aware manner.
run_async_subprocess(cmd)Invokes an async subprocess to run a command.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job handle to database
set_job_status(status)Validates and sets the job state
setup(task_metadata)Executor specific setup method
submit_task(submit_metadata)Abstract method that invokes the task on the remote backend.
teardown(task_metadata)Executor specific teardown method
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- boto_session_options()#
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
- Return type
Dict[str,str]
- async cancel()[source]#
Abstract method that sends a cancellation request to the remote backend.
- Return type
bool
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- async get_cancel_requested()#
Get if the task was requested to be canceled
- Arg(s)
None
- Return(s)
Whether the task has been requested to be cancelled
- Return type
Any
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- async get_status(braket, job_arn)[source]#
Query the status of a previously submitted Braket hybrid job.
- Parameters
braket – Braket client object.
job_arn (
str) – ARN used to identify a Braket hybrid job.
- Returns
String describing the job status.
- Return type
status
- async get_version_info()#
Query the database for dispatch version metadata.
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- async poll(task_group_metadata, data)#
Block until the job has reached a terminal state.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of send().
The return value of poll() will be passed directly into receive().
Raise NotImplementedError to indicate that the compute backend will notify the Covalent server asynchronously of job completion.
- Return type
Any
- async query_result(query_metadata)[source]#
Abstract method that retrieves the pickled result from the remote cache.
- Return type
Any
- async receive(task_group_metadata, data)#
Return a list of task updates.
Each task must have reached a terminal state by the time this is invoked.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of poll() or the request body of /jobs/update.
- Return type
List[TaskUpdate]- Returns
Returns a list of task results, each a TaskUpdate dataclass of the form
- {
“dispatch_id”: dispatch_id, “node_id”: node_id, “status”: status, “assets”: {
- ”output”: {
“remote_uri”: output_uri,
}, “stdout”: {
”remote_uri”: stdout_uri,
}, “stderr”: {
”remote_uri”: stderr_uri,
},
},
}
corresponding to the node ids (task_ids) specified in the task_group_metadata. This might be a subset of the node ids in the originally submitted task group as jobs may notify Covalent asynchronously of completed tasks before the entire task group finishes running.
- async run(function, args, kwargs, task_metadata)[source]#
Abstract method to run a function in the executor in async-aware manner.
- Parameters
function (
Callable) – The function to run in the executorargs (
List) – List of positional arguments to be used by the functionkwargs (
Dict) – Dictionary of keyword arguments to be used by the function.task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id
- Returns
The result of the function execution
- Return type
output
- async static run_async_subprocess(cmd)#
Invokes an async subprocess to run a command.
- Return type
Tuple
- async send(task_specs, resources, task_group_metadata)#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[TaskSpec]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- Return type
Any
- async set_job_handle(handle)#
Save the job handle to database
- Arg(s)
handle: JSONable type identifying the job being executed by the backend
- Return(s)
Response from the listener that handles inserting the job handle to database
- Return type
Any
- async set_job_status(status)#
Validates and sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- async setup(task_metadata)#
Executor specific setup method
- async submit_task(submit_metadata)[source]#
Abstract method that invokes the task on the remote backend.
- Parameters
task_metadata – Dictionary of metadata for the task. Current keys are dispatch_id and node_id.
- Returns
Task UUID defined on the remote backend.
- Return type
task_uuid
- async teardown(task_metadata)#
Executor specific teardown method
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- validate_status(status)#
Overridable filter
- Return type
bool
- async write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
This uses aiofiles to avoid blocking the event loop.
- Return type
None
AWS EC2 Executor#
Covalent is a Pythonic workflow tool used to execute tasks on advanced computing hardware.
This plugin allows tasks to be executed in an AWS EC2 instance (which is auto-created) when you execute your workflow with covalent.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-ec2-plugin
Note
Users will also need to have Terraform installed on their local machine in order to use this plugin.
This is a toy example of how a workflow can be adapted to utilize the EC2 Executor. Here we train a Support Vector Machine (SVM) and spin up an EC2 automatically to execute the train_svm electron. We also note we require DepsPip to install the dependencies on the EC2 instance.
from numpy.random import permutation
from sklearn import svm, datasets
import covalent as ct
deps_pip = ct.DepsPip(
packages=["numpy==1.23.2", "scikit-learn==1.1.2"]
)
executor = ct.executor.EC2Executor(
instance_type="t2.micro",
volume_size=8, #GiB
ssh_key_file="~/.ssh/ec2_key" # default key_name will be "ec2_key"
)
# Use executor plugin to train our SVM model.
@ct.electron(
executor=executor,
deps_pip=deps_pip
)
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
@ct.electron
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
@ct.electron
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],
y_test[:90]
)
@ct.lattice
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(
data=data,
clf=clf
)
return score
# Dispatch the workflow
dispatch_id = ct.dispatch(run_experiment)(
C=1.0,
gamma=0.7
)
# Wait for our result and get result value
result = ct.get_result(dispatch_id=dispatch_id, wait=True).result
print(result)
During the execution of the workflow one can navigate to the UI to see the status of the workflow, once completed however the above script should also output a value with the score of our model.
0.8666666666666667
Config Key |
Is Required |
Default |
Description |
|---|---|---|---|
profile |
No |
default |
Named AWS profile used for authentication |
region |
No |
us-east-1 |
AWS Region to use to for client calls to AWS |
credentials_file |
Yes |
~/.aws/credentials |
The path to the AWS credentials file |
ssh_key_file |
Yes |
~/.ssh/id_rsa |
The path to the private key that corresponds to the EC2 Key Pair |
instance_type |
Yes |
t2.micro |
The EC2 instance type that will be spun up automatically. |
key_name |
Yes |
Name of key specified in |
The name of the AWS EC2 Key Pair that will be used to SSH into EC2 instance |
volume_size |
No |
8 |
The size in GiB of the GP2 SSD disk to be provisioned with EC2 instance. |
vpc |
No |
(Auto created) |
The VPC ID that will be associated with the EC2 instance, if not specified a VPC will be created. |
subnet |
No |
(Auto created) |
The Subnet ID that will be associated with the EC2 instance, if not specified a public Subnet will be created. |
remote_cache |
No |
~/.cache/covalent |
The location on the EC2 instance where covalent artifacts will be created. |
This plugin can be configured in one of two ways:
Configuration options can be passed in as constructor keys to the executor class
ct.executor.EC2ExecutorBy modifying the covalent configuration file under the section
[executors.ec2]
The following shows an example of how a user might modify their covalent configuration file to support this plugin:
[executors.ec2]
ssh_key_file = "/home/user/.ssh/ssh_key.pem"
key_name = "ssh_key"
This plugin requires users have an AWS account. New users can follow instructions here to create a new account. In order to run workflows with Covalent and the AWS EC2 plugin, there are a few notable resources that need to be provisioned first. Whenever interacting with AWS resources, users strongly recommended to follow best practices for managing cloud credentials. Users are recommended to follow the principle of least privilege. For this executor, users who wish to deploy required infrastructure may use the AWS Managed Policy AmazonEC2FullAccess although some administrators may wish to further restrict instance families, regions, or other options according to their organization’s cloud policies.
The required resources include an EC2 Key Pair, and optionally a VPC & Subnet that can be used instead of the EC2 executor automatically creating it.
Resource |
Is Required |
Config Key |
Description |
|---|---|---|---|
AWS EC2 Key Pair |
Yes |
|
An EC2 Key Pair must be created and named corresponding to the |
VPC |
No |
|
A VPC ID can be provided corresponding to the |
Subnet |
No |
|
A Subnet ID can be provided corresponding to the |
Security Group |
No |
(Auto Created) |
A security group will be auto created and attached to the VPC in order to give the local machine (dispatching workflow) SSH access to the EC2 instance. |
EC2 Instance |
No |
(Auto Created) |
An EC2 Instance will be automatically provisioned for each electron in the workflow that utilizes this executor. |
To create an AWS EC2 Key pair refer to the following AWS documentation.
To create a VPC & Subnet refer to the following AWS documentation.
When tasks are run using this executor, the following infrastructure is ephemerally deployed.

This includes the minimal infrastructure needed to deploy an EC2 instance in a public subnet connected to an internet gateway. Users can validate that resources are correctly provisioned by monitoring the EC2 dashboard in the AWS Management Console. The overhead added by using this executor is on the order of several minutes, depending on the complexity of any additional user-specified runtime dependencies. Users are advised not to use any sensitive data with this executor without careful consideration of security policies. By default, data in transit is cached on the EBS volume attached to the EC2 instance in an unencrypted format.
These resources are torn down upon task completion and not shared across tasks in a workflow. Deployment of these resources will incur charges for EC2 alone; refer to AWS EC2 pricing for details. Note that this can be deployed in any AWS region in which the user is otherwise able to deploy EC2 instances. Some users may encounter quota limits when using EC2; this can be addressed by opening a support ticket with AWS.
AWS ECS Executor#
With this executor, users can execute tasks (electrons) or entire lattices using the AWS Elastic Container Service (ECS). This executor plugin is well suited for low to medium compute intensive electrons with modest memory requirements. Since AWS ECS offers very quick spin up times, this executor is a good fit for workflows with a large number of independent tasks that can be dispatched simultaneously.
1. Installation#
To use this plugin with Covalent, simply install it using pip:
pip install covalent-ecs-plugin
2. Usage Example#
This is an example of how a workflow can be constructed to use the AWS ECS executor. In the example, we join two words to form a phrase and return an excited phrase.
import covalent as ct
executor = ct.executor.ECSExecutor(
s3_bucket_name="covalent-fargate-task-resources",
ecr_repo_name="covalent-fargate-task-images",
ecs_cluster_name="covalent-fargate-cluster",
ecs_task_family_name="covalent-fargate-tasks",
ecs_task_execution_role_name="ecsTaskExecutionRole",
ecs_task_role_name="CovalentFargateTaskRole",
ecs_task_subnet_id="subnet-871545e1",
ecs_task_security_group_id="sg-0043541a",
ecs_task_log_group_name="covalent-fargate-task-logs",
vcpu=1,
memory=2,
poll_freq=10,
)
@ct.electron(executor=executor)
def join_words(a, b):
return ", ".join([a, b])
@ct.electron(executor=executor)
def excitement(a):
return f"{a}!"
@ct.lattice
def simple_workflow(a, b):
phrase = join_words(a, b)
return excitement(phrase)
dispatch_id = ct.dispatch(simple_workflow)("Hello", "World")
result = ct.get_result(dispatch_id, wait=True)
print(result)
During the execution of the workflow, one can navigate to the UI to see the status of the workflow. Once completed, the above script should also output the result:
Hello, World
In order for the above workflow to run successfully, one has to provision the required AWS resources as mentioned in 4. Required AWS Resources.
3. Overview of configuration#
The following table shows a list of all input arguments including the required arguments to be supplied when instantiating the executor:
Config Value |
Is Required |
Default |
Description |
|---|---|---|---|
credentials |
No |
~/.aws/credentials |
The path to the AWS credentials file |
profile |
No |
default |
The AWS profile used for authentication |
region |
Yes |
us-east-1 |
AWS region to use for client calls to AWS |
s3_bucket_name |
No |
covalent-fargate-task-resources |
The name of the S3 bucket where objects are stored |
ecr_repo_name |
No |
covalent-fargate-task-images |
The name of the ECR repository where task images are stored |
ecs_cluster_name |
No |
covalent-fargate-cluster |
The name of the ECS cluster on which tasks run |
ecs_task_family_name |
No |
covalent-fargate-tasks |
The name of the ECS task family for a user, project, or experiment. |
ecs_task_execution_role_name |
No |
CovalentFargateTaskRole |
The IAM role used by the ECS agent |
ecs_task_role_name |
No |
CovalentFargateTaskRole |
The IAM role used by the container during runtime |
ecs_task_subnet_id |
Yes |
Valid subnet ID |
|
ecs_task_security_group_id |
Yes |
Valid security group ID |
|
ecs_task_log_group_name |
No |
covalent-fargate-task-logs |
The name of the CloudWatch log group where container logs are stored |
vcpu |
No |
0.25 |
The number of vCPUs available to a task |
memory |
No |
0.5 |
The memory (in GB) available to a task |
poll_freq |
No |
10 |
The frequency (in seconds) with which to poll a submitted task |
cache_dir |
No |
/tmp/covalent |
The cache directory used by the executor for storing temporary files |
The following snippet shows how users may modify their Covalent configuration to provide the necessary input arguments to the executor:
[executors.ecs]
credentials = "~/.aws/credentials"
profile = "default"
s3_bucket_name = "covalent-fargate-task-resources"
ecr_repo_name = "covalent-fargate-task-images"
ecs_cluster_name = "covalent-fargate-cluster"
ecs_task_family_name = "covalent-fargate-tasks"
ecs_task_execution_role_name = "ecsTaskExecutionRole"
ecs_task_role_name = "CovalentFargateTaskRole"
ecs_task_subnet_id = "<my-subnet-id>"
ecs_task_security_group_id = "<my-security-group-id>"
ecs_task_log_group_name = "covalent-fargate-task-logs"
vcpu = 0.25
memory = 0.5
cache_dir = "/tmp/covalent"
poll_freq = 10
Within a workflow, users can use this executor with the default values configured in the configuration file as follows:
import covalent as ct
@ct.electron(executor="ecs")
def task(x, y):
return x + y
Alternatively, users can customize this executor entirely by providing their own values to its constructor as follows:
import covalent as ct
from covalent.executor import ECSExecutor
ecs_executor = ECSExecutor(
credentials="my_custom_credentials",
profile="my_custom_profile",
s3_bucket_name="my_s3_bucket",
ecr_repo_name="my_ecr_repo",
ecs_cluster_name="my_ecs_cluster",
ecs_task_family_name="my_custom_task_family",
ecs_task_execution_role_name="myCustomTaskExecutionRole",
ecs_task_role_name="myCustomTaskRole",
ecs_task_subnet_id="my-subnet-id",
ecs_task_security_group_id="my-security-group-id",
ecs_task_log_group_name="my-task-log-group",
vcpu=1,
memory=2,
cache_dir="/home/<user>/covalent/cache",
poll_freq=10,
)
@ct.electron(executor=ecs_executor)
def task(x, y):
return x + y
4. Required AWS Resources#
This executor uses different AWS services (S3, ECR, ECS, and Fargate) to successfully run a task. In order for the executor to work end-to-end, the following resources need to be configured either with Terraform or manually provisioned on the AWS Dashboard
Resource |
Config Name |
Description |
|---|---|---|
IAM Role |
ecs_task_execution_role_name |
The IAM role used by the ECS agent |
IAM Role |
ecs_task_role_name |
The IAM role used by the container during runtime |
S3 Bucket |
s3_bucket_name |
The name of the S3 bucket where objects are stored |
ECR repository |
ecr_repo_name |
The name of the ECR repository where task images are stored |
ECS Cluster |
ecs_cluster_name |
The name of the ECS cluster on which tasks run |
ECS Task Family |
ecs_task_family_name |
The name of the task family that specifies container information for a user, project, or experiment |
VPC Subnet |
ecs_task_subnet_id |
The ID of the subnet where instances are created |
Security group |
ecs_task_security_group_id |
The ID of the security group for task instances |
Cloudwatch log group |
ecs_task_log_group_name |
The name of the CloudWatch log group where container logs are stored |
CPU |
vCPU |
The number of vCPUs available to a task |
Memory |
memory |
The memory (in GB) available to a task |
The following IAM roles and policies must be properly configured so that the executor has all the necessary permissions to interact with the different AWS services:
ecs_task_execution_role_nameis the IAM role used by the ECS agentecs_task_role_nameis the IAM role used by the container during runtime
If omitted, these IAM role names default to ecsTaskExecutionRole and CovalentFargateTaskRole, respectively.
The IAM policy attached to the ecsTaskExecutionRole is the following:
ECS Task Execution Role IAM Policy
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"ecr:GetAuthorizationToken",
"ecr:BatchCheckLayerAvailability",
"ecr:GetDownloadUrlForLayer",
"ecr:BatchGetImage",
"logs:CreateLogStream",
"logs:PutLogEvents"
],
"Resource": "*"
}
]
}
These policies allow the service to download container images from ECR so that the tasks can be executed on an ECS
cluster. The policy attached to the CovalentFargateTaskRole is as follows
AWS Fargate Task Role IAM Policy
{
"Version": "2012-10-17",
"Statement": [
{
"Sid": "VisualEditor0",
"Effect": "Allow",
"Action": "braket:*",
"Resource": "*"
},
{
"Sid": "VisualEditor1",
"Effect": "Allow",
"Action": [
"s3:PutObject",
"s3:GetObject",
"s3:ListBucket"
],
"Resource": [
"arn:aws:s3:::covalent-fargate-task-resources/*",
"arn:aws:s3:::covalent-fargate-task-resources"
]
}
]
}
Users can provide their custom IAM roles/policies as long as they respect the permissions listed in the above documents. For more information on how to create IAM roles and attach policies in AWS, refer to IAM roles.
The executor also requires a proper networking setup so that the containers can be properly launched into their respective
subnets. The executor requires that the user provide a subnet ID and a security group ID prior to using the executor
in a workflow.
The executor uses Docker to build container images with the task function code baked into the image. The resulting image is pushed into the elastic container registry provided by the user. Following this, an ECS task definition using the user provided arguments is registered and the corresponding task container is launched. The output from the task is uploaded to the S3 bucket provided by the user and parsed to obtain the result object. In order for the executor to properly run and build images, users must have Docker installed and properly configured on their machines.
AWS Lambda Executor#
With this executor, users can execute tasks (electrons) or entire lattices using the AWS Lambda serverless compute service. It is appropriate to use this plugin for electrons that are expected to be short lived, low in compute intensity. This plugin can also be used for workflows with a high number of electrons that are embarassingly parallel (fully independent of each other).
The following AWS resources are required by this executor
Container based AWS Lambda function
AWS S3 bucket for caching objects
IAM role for Lambda
ECR container registry for storing docker images
1. Installation#
To use this plugin with Covalent, simply install it using pip:
pip install covalent-awslambda-plugin
Note
Due to the isolated nature of AWS Lambda, the packages available on that environment are limited. This means that only the modules that come with python out-of-the-box are accessible to your function. Deps are also limited in a similar fashion. However, AWS does provide a workaround for pip package installations: https://aws.amazon.com/premiumsupport/knowledge-center/lambda-python-package-compatible/.
2. Usage Example#
This is an example of how a workflow can be constructed to use the AWS Lambda executor. In the example, we join two words to form a phrase and return an excited phrase.
import covalent as ct
from covalent.executor import AWSLambdaExecutor
executor = AWSLambdaExecutor(
function_name = "my-lambda-function"
s3_bucket_name="covalent-lambda-job-resources"
)
@ct.electron(executor=executor)
def join_words(a, b):
return ",".join([a, b])
@ct.electron(executor=executor)
def excitement(a):
return f"{a}!"
@ct.lattice
def simple_workflow(a, b):
phrase = join_words(a, b)
return excitement(phrase)
dispatch_id = ct.dispatch(simple_workflow)("Hello", "World")
result = ct.get_result(dispatch_id, wait=True)
print(result)
During the execution of the workflow, one can navigate to the UI to see the status of the workflow. Once completed, the above script should also output the result:
Hello, World!
In order for the above workflow to run successfully, one has to provision the required AWS resources as mentioned in 4. Required AWS Resources.
Note
Users may encounter failures with dispatching workflows on MacOS due to errors with importing the psutil module. This is a known issue and will be addressed in a future sprint.
3. Overview of configuration#
The following table shows a list of all input arguments including the required arguments to be supplied when instantiating the executor:
Config Value |
Is Required |
Default |
Description |
|---|---|---|---|
function_name |
Yes |
|
Name of the AWS lambda function to be used at runtime |
s3_bucket_name |
Yes |
|
Name of an AWS S3 bucket that the executor must use to cache object files |
credentials_file |
No |
~/.aws/credentials |
The path to your AWS credentials file |
profile |
No |
default |
AWS profile used for authentication |
poll_freq |
No |
5 |
Time interval between successive polls to the lambda function |
cache_dir |
No |
~/.cache/covalent |
Path on the local file system to a cache |
timeout |
No |
|
Duration in seconds to keep polling the task for results/exceptions raised |
The following snippet shows how users may modify their Covalent configuration to provide the necessary input arguments to the executor:
[executors.awslambda]
function_name = "my-lambda-function"
s3_bucket_name = "covalent-lambda-job-resources"
credentials_file = "/home/<user>/.aws/credentials"
profile = "default"
region = "us-east-1"
cache_dir = "/home/<user>/.cache/covalent"
poll_freq = 5
timeout = 60
Within a workflow, users can use this executor with the default values configured in the configuration file as follows:
import covalent as ct
@ct.electron(executor="awslambda")
def task(x, y):
return x + y
Alternatively, users can customize this executor entirely by providing their own values to its constructor as follows:
import covalent as ct
from covalent.executor import AWSLambdaExecutor
lambda_executor = AWSLambdaExecutor(
function_name = "my-lambda-function"
s3_bucket_name="my_s3_bucket",
credentials_file="my_custom_credentials",
profile="custom_profile",
region="us-east-1",
cache_dir="/home/<user>/covalent/cache",
poll_freq=5,
timeout=60
)
@ct.electron(executor=lambda_executor)
def task(x, y):
return x + y
4. Required AWS Resources#
In order for the executor to work end-to-end, the following resources need to be provisioned apriori.
Resource |
Config Name |
Description |
|---|---|---|
IAM Role |
lambda_role_name |
The IAM role this lambda will assume during execution of your tasks |
S3 Bucket |
s3_bucket_name |
Name of an AWS S3 bucket that the executor can use to store temporary files |
AWS Lambda function |
function_name |
Name of the AWS lambda function created in AWS |
AWS Elastic Container Registry (ECR) |
|
The container registry that contains the docker images used by the lambda function to execute tasks |
The following JSON policy document shows the necessary IAM permissions required for the executor to properly run tasks using the AWS Lambda compute service:
IAM Policy
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"s3:*",
"s3-object-lambda:*"
],
"Resource": [
"arn:aws:s3:::<bucket-name>",
"arn:aws:s3:::<bucket-name>/*"
]
},
{
"Effect": "Allow",
"Action": [
"cloudformation:DescribeStacks",
"cloudformation:ListStackResources",
"cloudwatch:ListMetrics",
"cloudwatch:GetMetricData",
"ec2:DescribeSecurityGroups",
"ec2:DescribeSubnets",
"ec2:DescribeVpcs",
"kms:ListAliases",
"iam:GetPolicy",
"iam:GetPolicyVersion",
"iam:GetRole",
"iam:GetRolePolicy",
"iam:ListAttachedRolePolicies",
"iam:ListRolePolicies",
"iam:ListRoles",
"lambda:*",
"logs:DescribeLogGroups",
"states:DescribeStateMachine",
"states:ListStateMachines",
"tag:GetResources",
"xray:GetTraceSummaries",
"xray:BatchGetTraces"
],
"Resource": "*"
},
{
"Effect": "Allow",
"Action": "iam:PassRole",
"Resource": "*",
"Condition": {
"StringEquals": {
"iam:PassedToService": "lambda.amazonaws.com"
}
}
},
{
"Effect": "Allow",
"Action": [
"logs:DescribeLogStreams",
"logs:GetLogEvents",
"logs:FilterLogEvents"
],
"Resource": "arn:aws:logs:*:*:log-group:/aws/lambda/*"
}
]
}
where <bucket-name> is the name of an S3 bucket to be used by the executor to store temporary files generated during task
execution. The lambda function interacts with the S3 bucket as well as with the AWS Cloudwatch service to route any log messages.
Due to this, the lambda function must have the necessary IAM permissions in order to do so. Users must provision an IAM role that has
the AWSLambdaExecute policy attached to it. The policy document is summarized here for convenience:
Covalent Lambda Execution Role Policy
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"logs:*"
],
"Resource": "arn:aws:logs:*:*:*"
},
{
"Effect": "Allow",
"Action": [
"s3:GetObject",
"s3:PutObject"
],
"Resource": "arn:aws:s3:::*"
}
]
}
Users can use the following Terraform snippet as a starting point to spin up the required resources
provider aws {}
resource aws_s3_bucket bucket {
bucket = "my-s3-bucket"
}
resource aws_iam_role lambda_iam {
name = var.aws_lambda_iam_role_name
assume_role_policy = jsonencode({
Version = "2012-10-17"
Statement = [
{
Action = "sts:AssumeRole"
Effect = "Allow"
Sid = ""
Principal = {
Service = "lambda.amazonaws.com"
}
},
]
})
managed_policy_arns = [ "arn:aws:iam::aws:policy/AWSLambdaExecute" ]
}
resource aws_ecr_repository lambda_ecr {
name = "lambda_container_registry"
}
resource aws_lambda_function lambda {
function_name = "my-lambda-function"
role = aws_iam_role.lambda_iam.arn
packge_type = "Image"
timeout = <timeout value in seconds, max 900 (15 minutes), defaults to 3>
memory_size = <Max memory in MB that the Lambda is expected to use, defaults to 128>
image_uri = aws_ecr_repository.lambda_ecr.repository_url
}
For more information on how to create IAM roles and attach policies in AWS, refer to IAM roles. For more information on AWS S3, refer to AWS S3.
Note
The lambda function created requires a docker image to execute the any tasks required by it. We distribute ready to use AWS Lambda executor docker images that user’s can pull and push to their private ECR registries before dispatching workflows.
The base docker image can be obtained as follows
docker pull public.ecr.aws/covalent/covalent-lambda-executor:stable
Once the image has been obtained, user’s can tag it with their registry information and upload to ECR as follows
aws ecr get-login-password --region <region> | docker login --username AWS --password-stdin <aws_account_id>.dkr.ecr.<region>.amazonaws.com
docker tag public.ecr.aws/covalent/covalent-lambda-executor:stable <aws_account_id>.dkr.ecr.<region>.amazonaws.com/<my-repository>:tag
docker push <aws_account_id>.dkr.ecr.<region>.amazonaws.com/<my-repository>:tag
5. Custom Docker images#
As mentioned earlier, the AWS Lambda executor uses a docker image to execute an electron from a workflow. We distribute AWS Lambda executor base docker images that contain just the essential dependencies such as covalent and covalent-aws-plugins. However if the electron to be executed using the Lambda executor depends on Python packages that are not present in the base image by default, users will have to a build custom images prior to running their Covalent workflows using the AWS Lambda executor. In this section we cover the necessary steps required to extend the base executor image by installing additional Python packages and pushing the derived image to a private elastic container registry (ECR)
Note
Using PipDeps as described in the ../deps section with the AWS Lambda executor is currently not supported as it modifies the execution environment of the lambda function at runtime. As per AWS best practices for Lambda it is recommended to ship the lambda function as a self-contained object that has all of its dependencies in a deployment package/container image as described in detail here
All of our base AWS executor images are available in the AWS public registries and can be downloaded locally for consumption as described here here. For instance the stable AWS Lambda executor image can be downloaded from public ECR as follows
aws ecr-public get-login-password --region <aws-region> | docker login --username AWS --password-stdin public.ecr.aws
docker pull public.ecr.aws/covalent/covalent-lambda-executor:stable
Note
Executor images with the latest tag are also routinely pushed to the same registry. However, we strongly recommended using the stable tag when running executing workflows usin the AWS Lambda executor. The <aws-region> is a placeholder for the actual AWS region to be used by the user
Once the lambda base executor image has been downloaded, users can build upon that image by installing all the Python packages required by their tasks. The base executor uses a build time argument named LAMBDA_TASK_ROOT to set the install path of all python packages to /var/task inside the image. When extending the base image by installing additional python packages, it is recommended to install them to the same location so that they get resolved properly during runtime. Following is a simple example of how users can extend the AWS lambda base image by creating their own Dockerfile and installting additional packages such as numpy, pandas and scipy.
# Dockerfile
FROM public.ecr.aws/covalent/covalent-lambda-executor:stable as base
RUN pip install --target ${LAMBDA_TASK_ROOT} numpy pandas scipy
Warning
Do not override the entrypoint of the base image in the derived image when installing new packages. The docker ENTRYPOINT of the base image is what that gets trigged when AWS invokes your lambda function to execute the workflow electron
Once the Dockerfile has been created the derived image can be built as follows
docker build -f Dockerfile -t my-custom-lambda-executor:latest
After a successful build of the derived image, it needs to be uploaded to ECR so that it can be consumed by a lambda function when triggered by Covalent. As as first step, it is required to create an elastic container registry to hold the dervied executor images. This can be easily done by using the AWS CLI tool as follows
aws ecr create-repository --region <aws-region> --repository-name covalent/my-custom-lambda-executor
To upload the derived image to this registry, we would need to tag our local image as per the AWS guide and push the image to the registry as described here. To push an image, first one needs to authenticate with AWS and login their docker client
aws ecr get-login-password --region <aws-region> | docker login --username AWS --password-stdin <aws-account-id>.dkr.ecr.region.amazonaws.com
Once the login is successful, the local image needs to be re-tagged with the ECR repository information. If the image tag is omitted, latest is applied by default. In the following code block we show how to tag the derived image my-custom-lambda-executor:latest with the ECR information so that it can be uploaded successfully
docker tag my-custom-lambda-executor:latest <aws-account-id>.dkr.ecr.<aws-region>.amazonaws.com/my-custom-lambda-executor:latest
Note
<aws-account-id> and <aws-region> are placeholders for the actual AWS account ID and region to be used by the users
Once the derived image has been built and pushed to ECR, users need to create a Lambda function or update an existing one to use the new derived image instead of the base image executor image at runtime. A new AWS Lambda function can be quite easily created using the AWS Lambda CLI create-function command as follows
aws lambda create-function --function-name "my-covalent-lambda-function" --region <aws-region> \
--package-type Image \
--code ImageUri=<aws-account-id>.dkr.ecr.<aws-region>.amazonaws.com/my-custom-lambda-executor:latest \
--role <Lambda executor role ARN> \
--memory-size 512 \
--timeout 900
The above CLI command will register a new AWS lambda function that will use the user’s custom derived image my-custom-lambda-executor:latest with a memory size of 512 MB and a timeout values of 900 seconds. The role argument is used to specify the ARN of the IAM role the AWS Lambda can assume during execution. The necessary permissions for the IAM role have been provided in Required AWS resources section. More details about creating and updating AWS lambda functions can be found here.
- class covalent_awslambda_plugin.AWSLambdaExecutor(function_name=None, s3_bucket_name=None, credentials_file=None, profile=None, region=None, execution_role='', poll_freq=None, timeout=900)[source]#
AWS Lambda executor plugin
- Parameters
function_name (
Optional[str]) – Name of an existing lambda function to use during execution (default: covalent-awsambda-executor)s3_bucket_name (
Optional[str]) – Name of a AWS S3 bucket that the executor can use to store temporary files (default: covalent-lambda-job-resources)execution_role (
str) – Name of the IAM role assigned to the AWS Lambda functioncredentials_file (
Optional[str]) – Path to AWS credentials file (default: ~/.aws/credentials)profile (
Optional[str]) – AWS profile (default: default)region (
Optional[str]) – AWS region (default: us-east-1)poll_freq (
Optional[int]) – Time interval between successive polls to the lambda function (default: 5)timeout (
int) – Duration in seconds to poll Lambda function for results (default: 900)
Methods:
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
cancel()Cancel execution
from_dict(object_dict)Rehydrate a dictionary representation
Get if the task was requested to be canceled
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
Yield a boto3 session to be used for instantiating AWS service clients/resources
get_status(object_key)Return status of availability of result object on remote machine
Query the database for dispatch version metadata.
poll(task_group_metadata, data)Block until the job has reached a terminal state.
query_result(workdir, result_filename)Abstract method that retrieves the pickled result from the remote cache.
query_result_sync(workdir, result_filename)Fetch the result object from the S3 bucket
query_task_exception_sync(workdir, …)Fetch the exception raised from the S3 bucket
receive(task_group_metadata, data)Return a list of task updates.
run(function, args, kwargs, task_metadata)Run the executor
run_async_subprocess(cmd)Invokes an async subprocess to run a command.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job handle to database
set_job_status(status)Validates and sets the job state
setup(task_metadata)Executor specific setup method
submit_task(function_name, func_filename, …)Submit the task by invoking the AWS Lambda function
submit_task_sync(function_name, …)The actual (blocking) submit_task function
teardown(task_metadata)Executor specific teardown method
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- boto_session_options()#
Returns a dictionary of kwargs to populate a new boto3.Session() instance with proper auth, region, and profile options.
- Return type
Dict[str,str]
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- async get_cancel_requested()#
Get if the task was requested to be canceled
- Arg(s)
None
- Return(s)
Whether the task has been requested to be cancelled
- Return type
Any
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- get_session()[source]#
Yield a boto3 session to be used for instantiating AWS service clients/resources
- Parameters
None –
- Returns
AWS boto3.Session object
- Return type
session
- async get_status(object_key)[source]#
Return status of availability of result object on remote machine
- Parameters
object_key (
str) – Name of the S3 object- Returns
bool indicating whether the object exists or not on S3 bucket
- async get_version_info()#
Query the database for dispatch version metadata.
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- async poll(task_group_metadata, data)#
Block until the job has reached a terminal state.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of send().
The return value of poll() will be passed directly into receive().
Raise NotImplementedError to indicate that the compute backend will notify the Covalent server asynchronously of job completion.
- Return type
Any
- async query_result(workdir, result_filename)[source]#
Abstract method that retrieves the pickled result from the remote cache.
- query_result_sync(workdir, result_filename)[source]#
Fetch the result object from the S3 bucket
- Parameters
workdir (
str) – Path on the local file system where the pickled object is downloaded- Returns
None
- query_task_exception_sync(workdir, exception_filename)[source]#
Fetch the exception raised from the S3 bucket
- Parameters
workdir (
str) – Path on the local file system where the exception json dump is downloaded- Returns
None
- async receive(task_group_metadata, data)#
Return a list of task updates.
Each task must have reached a terminal state by the time this is invoked.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of poll() or the request body of /jobs/update.
- Return type
List[TaskUpdate]- Returns
Returns a list of task results, each a TaskUpdate dataclass of the form
- {
“dispatch_id”: dispatch_id, “node_id”: node_id, “status”: status, “assets”: {
- ”output”: {
“remote_uri”: output_uri,
}, “stdout”: {
”remote_uri”: stdout_uri,
}, “stderr”: {
”remote_uri”: stderr_uri,
},
},
}
corresponding to the node ids (task_ids) specified in the task_group_metadata. This might be a subset of the node ids in the originally submitted task group as jobs may notify Covalent asynchronously of completed tasks before the entire task group finishes running.
- async run(function, args, kwargs, task_metadata)[source]#
Run the executor
- Parameters
function (
Callable) – Python callable to be executed on the remote executorargs (
List) – List of positional arguments to be passed to the functionkwargs (
Dict) – Keyword arguments to be passed into the functiontask_metadata (
Dict) – Dictionary containing the task dispatch_id and node_id
- Returns
None
- async static run_async_subprocess(cmd)[source]#
Invokes an async subprocess to run a command.
- Return type
Tuple
- async send(task_specs, resources, task_group_metadata)#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[TaskSpec]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- Return type
Any
- async set_job_handle(handle)#
Save the job handle to database
- Arg(s)
handle: JSONable type identifying the job being executed by the backend
- Return(s)
Response from the listener that handles inserting the job handle to database
- Return type
Any
- async set_job_status(status)#
Validates and sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- async setup(task_metadata)#
Executor specific setup method
- async submit_task(function_name, func_filename, result_filename, exception_filename)[source]#
Submit the task by invoking the AWS Lambda function
- Parameters
function_name (
str) – AWS Lambda function name- Returns
AWS boto3 client invoke lambda response
- Return type
response
- submit_task_sync(function_name, func_filename, result_filename, exception_filename)[source]#
The actual (blocking) submit_task function
- Return type
Dict
- async teardown(task_metadata)#
Executor specific teardown method
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- validate_status(status)#
Overridable filter
- Return type
bool
- async write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
This uses aiofiles to avoid blocking the event loop.
- Return type
None
Azure Batch Executor#
Covalent Azure Batch executor is an interface between Covalent and Microsoft Azure Batch. This executor allows execution of Covalent tasks on Azure’s Batch service.
The batch executor is well suited for compute/memory intensive tasks since the resource pool of compute virtual machines can be scaled accordingly. Furthermore, Azure Batch allows running tasks in parallel on multiple virtual machines and their scheduling engine manages execution of the tasks.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-azurebatch-plugin
In this example, we train a Support Vector Machine (SVM) using an instance of the Azure Batch executor. The train_svm electron is submitted as a batch job in an existing Azure Batch Compute environment. Note that we also require DepsPip in order to install the python package dependencies before executing the electron in the batch environment.
from numpy.random import permutation
from sklearn import svm, datasets
import covalent as ct
from covalent.executor import AzureBatchExecutor
deps_pip = ct.DepsPip(
packages=["numpy==1.22.4", "scikit-learn==1.1.2"]
)
executor = AzureBatchExecutor(
tenant_id="tenant-id",
client_id="client-id",
client_secret="client-secret",
batch_account_url="https://covalent.eastus.batch.azure.com",
batch_account_domain="batch.core.windows.net",
storage_account_name="covalentbatch",
storage_account_domain="blob.core.windows.net",
base_image_uri="covalent.azurecr.io/covalent-executor-base:latest",
pool_id="covalent-pool",
retries=3,
time_limit=300,
cache_dir="/tmp/covalent",
poll_freq=10
)
# Use executor plugin to train our SVM model
@ct.electron(
executor=executor,
deps_pip=deps_pip
)
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
@ct.electron
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
@ct.electron
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],y_test[:90]
)
@ct.lattice
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(
data=data,
clf=clf
)
return score
# Dispatch the workflow.
dispatch_id = ct.dispatch(run_experiment)(
C=1.0,
gamma=0.7
)
# Wait for our result and get result value
result = ct.get_result(dispatch_id, wait=True).result
print(result)
During the execution of the workflow, one can navigate to the UI to see the status of the workflow. Once completed, the above script should also output a value with the score of our model.
0.8666666666666667
Config Key |
Required |
Default |
Description |
|---|---|---|---|
tenant_id |
Yes |
None |
Azure tenant ID |
client_id |
Yes |
None |
Azure client ID |
client_secret |
Yes |
None |
Azure client secret |
batch_account_url |
Yes |
None |
Azure Batch account URL |
batch_account_domain |
No |
batch.core.windows.net |
Azure Batch account domain |
storage_account_name |
Yes |
None |
Azure Storage account name |
storage_account_domain |
No |
blob.core.windows.net |
Azure Storage account domain |
base_image_uri |
No |
covalent.azurecr.io/covalent-executor-base:latest |
Image used to run Covalent tasks |
pool_id |
Yes |
None |
Azure Batch pool ID |
retries |
No |
3 |
Number of retries for Azure Batch job |
time_limit |
No |
300 |
Time limit for Azure Batch job |
cache_dir |
No |
/tmp/covalent |
Directory to store cached files |
poll_freq |
No |
10 |
Polling frequency for Azure Batch job |
Configuration options can be passed in as constructor keys to the executor class
ct.executor.AzureBatchExecutorBy modifying the covalent configuration file under the section
[executors.azurebatch]
The following shows an example of how a user might modify their covalent configuration file to support this plugin:
[executors.azurebatch]
tenant_id="tenant-id",
client_id="client-id",
client_secret="client-secret",
batch_account_url="https://covalent.eastus.batch.azure.com",
batch_account_domain="batch.core.windows.net",
storage_account_name="covalentbatch",
storage_account_domain="blob.core.windows.net",
base_image_uri="my-custom-base-image",
pool_id="covalent-pool",
retries=5,
time_limit=500,
...
In some cases, users may wish to specify a custom base image for Covalent tasks running on Azure Batch. For instance, some orgazations may have pre-built environments containing application runtimes that may be otherwise difficult to configure at runtime. Similarly, some packages may be simple to install but greatly increase the memory and runtime overhead for a task. In both of these scenarios, custom containers can simplify the user experience.
To incorporate a custom container that can be used by Covalent tasks on Azure Batch, first locate the Dockerfile packaged with this plugin in covalent_azurebatch_plugin/assets/infra/Dockerfile. Assuming the custom container already has a compatible version of Python installed (specifically, the same version used by the Covalent SDK), build this image using the command
# Login to ACR registry first
acr login --name=<my_custom_registry_name>
# Build the combined image used by tasks
docker build --build-arg COVALENT_BASE_IMAGE=<my_custom_image_uri> -t <my_custom_registry_name>.azurecr.io/<my_custom_image_name>:latest .
# Push to the registry
docker push <my_custom_registry_name>.azurecr.io/<my_custom_image_name>:latest
where my_custom_image_uri is the fully qualified URI to the user’s image, my_custom_registry_name is the name of the ACR resource created during deployment of the resources below, and my_custom_image_name is the name of the output which contains both Covalent and the user’s custom image dependencies. Users would then use base_image_name=<my_custom_registry_name>.azurecr.io/<my_custom_image_name>:latest in the Azure Batch executor or associated configuration.
In order to use this plugin, the following Azure resources need to be provisioned first. These resources can be created using the Azure Portal or the Azure CLI.
Resource |
Is Required |
Config Key |
Description |
|---|---|---|---|
Batch Account |
Yes |
|
A batch account is required to submit jobs to Azure Batch. The URL can be found under the Account endpoint field in the Batch account. Furthermore, ensure that |
Storage Account |
Yes |
|
Storage account must be created with blob service enabled in order for covalent to store essential files that are needed during execution. |
Resource Group |
Yes |
N/A |
The resource group is a logical grouping of Azure resources that can be managed as one entity in terms of lifecycle and security. |
Container Registry |
Yes |
N/A |
Container registry is required to store any custom containers used to run Batch jobs. |
Pool ID |
Yes |
|
A pool is a collection of compute nodes that are managed together. The pool ID is the name of the pool that will be used to execute the jobs. |
More information on authentication with service principals and necessary permissions for this executor can be found here.
For more information on error handling and detection in Batch, refer to the Microsoft Azure documentation. Furthermore, information on best practices can be found here.
Google Batch Executor#

Covalent Google Batch executor is an interface between Covalent and Google Cloud Platform’s Batch compute service. This executor allows execution of Covalent tasks on Google Batch compute service.
This batch executor is well suited for tasks with high compute/memory requirements. The compute resources required can be very easily configured/specified in the executor’s configuration. Google Batch scales really well thus allowing users to queue and execute multiple tasks concurrently on their resources efficiently. Google’s Batch job scheduler manages the complexity of allocating the resources needed by the task and de-allocating them once the job has finished.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-gcpbatch-plugin
Here we present an example on how a user can use the GCP Batch executor plugin in their Covalent workflows. In this example we train a simple SVM (support vector machine) model using the Google Batch executor. This executor is quite minimal in terms of the required cloud resources that need to be provisioned prior to first use. The Google Batch executor needs the following cloud resources pre-configured:
A Google storage bucket
Cloud artifact registry for Docker images
- A service account with the following permissions
roles/batch.agentReporterroles/logging.logWriterroles/logging.viewerroles/artifactregistry.readerroles/storage.objectCreatorroles/storage.objectViewer
Note
Details about Google services accounts and how to use them properly can be found here.
from numpy.random import permutation
from sklearn import svm, datasets
import covalent as ct
deps_pip = ct.DepsPip(
packages=["numpy==1.23.2", "scikit-learn==1.1.2"]
)
executor = ct.executor.GCPBatchExecutor(
bucket_name = "my-gcp-bucket",
region='us-east1',
project_id = "my-gcp-project-id",
container_image_uri = "my-executor-container-image-uri",
service_account_email = "my-service-account-email",
vcpus = 2, # Number of vCPUs to allocate
memory = 512, # Memory in MB to allocate
time_limit = 300, # Time limit of job in seconds
poll_freq = 3 # Number of seconds to pause before polling for the job's status
)
# Use executor plugin to train our SVM model.
@ct.electron(
executor=executor,
deps_pip=deps_pip
)
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
@ct.electron
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
@ct.electron
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],
y_test[:90]
)
@ct.lattice
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(
data=data,
clf=clf
)
return score
# Dispatch the workflow
dispatch_id = ct.dispatch(run_experiment)(
C=1.0,
gamma=0.7
)
# Wait for our result and get result value
result = ct.get_result(dispatch_id=dispatch_id, wait=True).result
print(result)
During the execution of the workflow the user can navigate to the web-based browser UI to see the status of the computations.
Config Key |
Is Required |
Default |
Description |
|---|---|---|---|
project_id |
yes |
None |
Google cloud project ID |
region |
No |
us-east1 |
Google cloud region to use to for submitting batch jobs |
bucket_name |
Yes |
None |
Name of the Google storage bucket to use for storing temporary objects |
container_image_uri |
Yes |
None |
GCP Batch executor base docker image uri |
service_account_email |
Yes |
None |
Google service account email address that is to be used by the batch job when interacting with the resources |
vcpus |
No |
2 |
Number of vCPUs needed for the task. |
memory |
No |
256 |
Memory (in MB) needed by the task. |
retries |
No |
3 |
Number of times a job is retried if it fails. |
time_limit |
No |
300 |
Time limit (in seconds) after which jobs are killed. |
poll_freq |
No |
5 |
Frequency (in seconds) with which to poll a submitted task. |
cache_dir |
No |
/tmp/covalent |
Cache directory used by this executor for temporary files. |
This plugin can be configured in one of two ways:
Configuration options can be passed in as constructor keys to the executor class
ct.executor.GCPBatchExecutorBy modifying the covalent configuration file under the section
[executors.gcpbatch]
[executors.gcpbatch]
bucket_name = <my-gcp-bucket-name>
project_id = <my-gcp-project-id>
container_image_uri = <my-base-executor-image-uri>
service_account_email = <my-service-account-email>
region = <google region for batch>
vcpus = 2 # number of vcpus needed by the job
memory = 256 # memory in MB required by the job
retries = 3 # number of times to retry the job if it fails
time_limit = 300 # time limit in seconds after which the job is to be considered failed
poll_freq = 3 # Frequency in seconds with which to poll the job for the result
cache_dir = "/tmp" # Path on file system to store temporary objects
In order to successfully execute tasks using the Google Batch executor, some cloud resources need to be provisioned apriori.
Google storage bucket
The executor uses a storage bucket to store/cache exception/result objects that get generated during execution.
Google Docker artifact registry
The executor submits a container job whose image is pulled from the provided container_image_uri argument of the executor.
Service account
Keeping good security practices in mind, the jobs are executed using a service account that only has the necessary permissions attached to it that are required for the job to finish.
Users can free to provision these resources as they see fit or they can use Covalent to provision these for them. Covalent CLI can be used to deploy the required cloud resources. Covalent behind the scenes uses Terraform to provision the cloud resources. The terraform HCL scripts can be found in the plugin’s Github repository here.
To run the scripts manually, users must first authenticate with Google cloud via their CLI and print out the access tokens with the following commands:
gcloud auth application-default login
gcloud auth print-access-token
Once the user has authenticated, the infrastructure can be deployed by running the Terraform commands in the infra folder of the plugin’s repository.
terraform plan -out tf.plan
terrafrom apply tf.plan -var="access_token=<access_token>"
Note
For first time deployment, the terraform provides must be initialized properly via terraform init.
The Terraform script also builds the base executor docker image and uploads it to the artifact registry after getting created. This means that users do not have to manually build and push the image.
Slurm Executor#
This executor plugin interfaces Covalent with HPC systems managed by Slurm. For workflows to be deployable, users must have SSH access to the Slurm login node, writable storage space on the remote filesystem, and permissions to submit jobs to Slurm.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-slurm-plugin
On the remote system, the Python version in the environment you plan to use must match that used when dispatching the calculations. Additionally, the remote system’s Python environment must have the base covalent package installed (e.g. pip install covalent).
The following shows an example of a Covalent configuration that is modified to support Slurm:
[executors.slurm]
username = "user"
address = "login.cluster.org"
ssh_key_file = "/home/user/.ssh/id_rsa"
remote_workdir = "/scratch/user"
cache_dir = "/tmp/covalent"
[executors.slurm.options]
nodes = 1
ntasks = 4
cpus-per-task = 8
constraint = "gpu"
gpus = 4
qos = "regular"
[executors.slurm.srun_options]
cpu_bind = "cores"
gpus = 4
gpu-bind = "single:1"
The first stanza describes default connection parameters for a user who can connect to the Slurm login node using, for example:
ssh -i /home/user/.ssh/id_rsa user@login.cluster.org
The second and third stanzas describe default parameters for #SBATCH directives and default parameters passed directly to srun, respectively.
This example generates a script containing the following preamble:
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=4
#SBATCH --cpus-per-task=8
#SBATCH --constraint=gpu
#SBATCH --gpus=4
#SBATCH --qos=regular
and subsequent workflow submission with:
srun --cpu_bind=cores --gpus=4 --gpu-bind=single:1
To use the configuration settings, an electron’s executor must be specified with a string argument, in this case:
import covalent as ct
@ct.electron(executor="slurm")
def my_task(x, y):
return x + y
Alternatively, passing a SlurmExecutor instance enables custom behavior scoped to specific tasks. Here, the executor’s prerun_commands and postrun_commands parameters can be used to list shell commands to be executed before and after submitting the workflow. These may include any additional srun commands apart from workflow submission. Commands can also be nested inside the submission call to srun by using the srun_append parameter.
More complex jobs can be crafted by using these optional parameters. For example, the instance below runs a job that accesses CPU and GPU resources on a single node, while profiling GPU usage via nsys and issuing complementary commands that pause/resume the central hardware counter.
executor = ct.executor.SlurmExecutor(
remote_workdir="/scratch/user/experiment1",
options={
"qos": "regular",
"time": "01:30:00",
"nodes": 1,
"constraint": "gpu",
},
prerun_commands=[
"module load package/1.2.3",
"srun --ntasks-per-node 1 dcgmi profile --pause"
],
srun_options={
"n": 4,
"c": 8,
"cpu-bind": "cores",
"G": 4,
"gpu-bind": "single:1"
},
srun_append="nsys profile --stats=true -t cuda --gpu-metrics-device=all",
postrun_commands=[
"srun --ntasks-per-node 1 dcgmi profile --resume",
]
)
@ct.electron(executor=executor)
def my_custom_task(x, y):
return x + y
Here the corresponding submit script contains the following commands:
module load package/1.2.3
srun --ntasks-per-node 1 dcgmi profile --pause
srun -n 4 -c 8 --cpu-bind=cores -G 4 --gpu-bind=single:1 \
nsys profile --stats=true -t cuda --gpu-metrics-device=all \
python /scratch/user/experiment1/workflow_script.py
srun --ntasks-per-node 1 dcgmi profile --resume
Some users may need two-factor authentication (2FA) to connect to a cluster. This plugin supports one form of 2FA using the sshproxy service developed by NERSC. When this plugin is configured to support sshproxy, the user’s SSH key and certificate will be refreshed automatically by Covalent if either it does not exist or it is expired. We assume that the user has already configured 2FA, used the sshproxy service on the command line without issue, and added the executable to their PATH. Note that this plugin assumes the script is called sshproxy, not sshproxy.sh. Further note that using sshproxy within Covalent is not required; a user can still run it manually and provide ssh_key_file and cert_file in the plugin constructor.
In order to enable sshproxy in this plugin, add the following block to your Covalent configuration while the server is stopped:
[executors.slurm.sshproxy]
hosts = [ "perlmutter-p1.nersc.gov" ]
password = "<password>"
secret = "<mfa_secret>"
For details on how to modify your Covalent configuration, refer to the documentation here.
Then, reinstall this plugin using pip install covalent-slurm-plugin[sshproxy] in order to pull in the oathtool package which will generate one-time passwords.
The hosts parameter is a list of hostnames for which the sshproxy service will be used. If the address provided in the plugin constructor is not present in this list, sshproxy will not be used. The password is the user’s password, not including the 6-digit OTP. The secret is the 2FA secret provided when a user registers a new device on Iris. Rather than scan the QR code into an authenticator app, inspect the Oath Seed URL for a string labeled secret=..., typically consisting of numbers and capital letters. Users can validate that correct OTP codes are being generated by using the command oathtool <secret> and using the 6-digit number returned in the “Test” option on the Iris 2FA page. Note that these values are stored in plaintext in the Covalent configuration file. If a user suspects credentials have been stolen or compromised, contact your systems administrator immediately to report the incident and request deactivation.
- class covalent_slurm_plugin.SlurmExecutor(username=None, address=None, ssh_key_file=None, cert_file=None, remote_workdir=None, slurm_path=None, conda_env=None, cache_dir=None, options=None, sshproxy=None, srun_options=None, srun_append=None, prerun_commands=None, postrun_commands=None, poll_freq=None, cleanup=None, **kwargs)[source]#
Slurm executor plugin class.
- Parameters
username (
Optional[str]) – Username used to authenticate over SSH.address (
Optional[str]) – Remote address or hostname of the Slurm login node.ssh_key_file (
Optional[str]) – Private RSA key used to authenticate over SSH.cert_file (
Optional[str]) – Certificate file used to authenticate over SSH, if required.remote_workdir (
Optional[str]) – Working directory on the remote cluster.slurm_path (
Optional[str]) – Path to the slurm commands if they are not found automatically.conda_env (
Optional[str]) – Name of conda environment on which to run the function.cache_dir (
Optional[str]) – Cache directory used by this executor for temporary files.options (
Optional[Dict]) – Dictionary of parameters used to build a Slurm submit script.srun_options (
Optional[Dict]) – Dictionary of parameters passed to srun inside submit script.srun_append (
Optional[str]) – Command nested into srun call.prerun_commands (
Optional[List[str]]) – List of shell commands to run before submitting with srun.postrun_commands (
Optional[List[str]]) – List of shell commands to run after submitting with srun.poll_freq (
Optional[int]) – Frequency with which to poll a submitted job.cleanup (
Optional[bool]) – Whether to perform cleanup or not on remote machine.
Methods:
cancel(task_metadata, job_handle)Method to cancel the job identified uniquely by the job_handle (base class)
from_dict(object_dict)Rehydrate a dictionary representation
Get if the task was requested to be canceled
get_dispatch_context(dispatch_info)Start a context manager that will be used to access the dispatch info for the executor.
get_status(info_dict, conn)Query the status of a job previously submitted to Slurm.
Query the database for dispatch version metadata.
perform_cleanup(conn, remote_func_filename, …)Function to perform cleanup on remote machine
poll(task_group_metadata, data)Block until the job has reached a terminal state.
receive(task_group_metadata, data)Return a list of task updates.
run(function, args, kwargs, task_metadata)Abstract method to run a function in the executor in async-aware manner.
send(task_specs, resources, task_group_metadata)Submit a list of task references to the compute backend.
set_job_handle(handle)Save the job handle to database
set_job_status(status)Validates and sets the job state
setup(task_metadata)Executor specific setup method
teardown(task_metadata)Executor specific teardown method
to_dict()Return a JSON-serializable dictionary representation of self
validate_status(status)Overridable filter
write_streams_to_file(stream_strings, …)Write the contents of stdout and stderr to respective files.
- async cancel(task_metadata, job_handle)#
Method to cancel the job identified uniquely by the job_handle (base class)
- Arg(s)
task_metadata: Metadata of the task to be cancelled job_handle: Unique ID of the job assigned by the backend
- Return(s)
False by default
- Return type
bool
- from_dict(object_dict)#
Rehydrate a dictionary representation
- Parameters
object_dict (
dict) – a dictionary representation returned by to_dict- Return type
- Returns
self
Instance attributes will be overwritten.
- async get_cancel_requested()#
Get if the task was requested to be canceled
- Arg(s)
None
- Return(s)
Whether the task has been requested to be cancelled
- Return type
Any
- get_dispatch_context(dispatch_info)#
Start a context manager that will be used to access the dispatch info for the executor.
- Parameters
dispatch_info (
DispatchInfo) – The dispatch info to be used inside current context.- Return type
AbstractContextManager[DispatchInfo]- Returns
A context manager object that handles the dispatch info.
- async get_status(info_dict, conn)[source]#
Query the status of a job previously submitted to Slurm.
- Parameters
info_dict (
dict) –a dictionary containing all necessary parameters needed to query the status of the execution. Required keys in the dictionary are:
A string mapping “job_id” to Slurm job ID.
- Returns
String describing the job status.
- Return type
status
- async get_version_info()#
Query the database for dispatch version metadata.
- Arg:
dispatch_id: Dispatch ID of the lattice
- Returns
python_version, “covalent”: covalent_version}
- Return type
{“python”
- async perform_cleanup(conn, remote_func_filename, remote_slurm_filename, remote_py_filename, remote_result_filename, remote_stdout_filename, remote_stderr_filename)[source]#
Function to perform cleanup on remote machine
- Parameters
remote_func_filename (
str) – Function file on remote machineremote_slurm_filename (
str) – Slurm script file on remote machineremote_py_filename (
str) – Python script file on remote machineremote_result_filename (
str) – Result file on remote machineremote_stdout_filename (
str) – Standard out file on remote machineremote_stderr_filename (
str) – Standard error file on remote machine
- Return type
None- Returns
None
- async poll(task_group_metadata, data)#
Block until the job has reached a terminal state.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of send().
The return value of poll() will be passed directly into receive().
Raise NotImplementedError to indicate that the compute backend will notify the Covalent server asynchronously of job completion.
- Return type
Any
- async receive(task_group_metadata, data)#
Return a list of task updates.
Each task must have reached a terminal state by the time this is invoked.
- Parameters
task_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.data (
Any) – The return value of poll() or the request body of /jobs/update.
- Return type
List[TaskUpdate]- Returns
Returns a list of task results, each a TaskUpdate dataclass of the form
- {
“dispatch_id”: dispatch_id, “node_id”: node_id, “status”: status, “assets”: {
- ”output”: {
“remote_uri”: output_uri,
}, “stdout”: {
”remote_uri”: stdout_uri,
}, “stderr”: {
”remote_uri”: stderr_uri,
},
},
}
corresponding to the node ids (task_ids) specified in the task_group_metadata. This might be a subset of the node ids in the originally submitted task group as jobs may notify Covalent asynchronously of completed tasks before the entire task group finishes running.
- async run(function, args, kwargs, task_metadata)[source]#
Abstract method to run a function in the executor in async-aware manner.
- Parameters
function (
Callable) – The function to run in the executorargs (
List) – List of positional arguments to be used by the functionkwargs (
Dict) – Dictionary of keyword arguments to be used by the function.task_metadata (
Dict) – Dictionary of metadata for the task. Current keys are dispatch_id and node_id
- Returns
The result of the function execution
- Return type
output
- async send(task_specs, resources, task_group_metadata)#
Submit a list of task references to the compute backend.
- Parameters
task_specs (
List[TaskSpec]) – a list of TaskSpecsresources (
ResourceMap) – a ResourceMap mapping task assets to URIstask_group_metadata (
Dict) – A dictionary of metadata for the task group. Current keys are dispatch_id, node_ids, and task_group_id.
The return value of send() will be passed directly into poll().
- Return type
Any
- async set_job_handle(handle)#
Save the job handle to database
- Arg(s)
handle: JSONable type identifying the job being executed by the backend
- Return(s)
Response from the listener that handles inserting the job handle to database
- Return type
Any
- async set_job_status(status)#
Validates and sets the job state
For use with send/receive API
- Return(s)
Whether the action succeeded
- Return type
bool
- async setup(task_metadata)#
Executor specific setup method
- to_dict()#
Return a JSON-serializable dictionary representation of self
- Return type
dict
- validate_status(status)#
Overridable filter
- Return type
bool
- async write_streams_to_file(stream_strings, filepaths, dispatch_id, results_dir)#
Write the contents of stdout and stderr to respective files.
- Parameters
stream_strings (
Iterable[str]) – The stream_strings to be written to files.filepaths (
Iterable[str]) – The filepaths to be used for writing the streams.dispatch_id (
str) – The ID of the dispatch which initiated the request.results_dir (
str) – The location of the results directory.
This uses aiofiles to avoid blocking the event loop.
- Return type
None
SSH Executor#
Executing tasks (electrons) via SSH in remote machine. This executor plugin interfaces Covalent with other machines accessible to the user over SSH. It is appropriate to use this plugin to distribute tasks to one or more compute backends which are not controlled by a cluster management system, such as computers on a LAN, or even a collection of small-form-factor Linux-based devices such as Raspberry Pis, NVIDIA Jetsons, or Xeon Phi co-processors.
To use this plugin with Covalent, simply install it using pip:
pip install covalent-ssh-plugin
The following shows an example of how a user might modify their Covalent configuration to support this plugin:
[executors.ssh]
username = "user"
hostname = "host.hostname.org"
remote_dir = "/home/user/.cache/covalent"
ssh_key_file = "/home/user/.ssh/id_rsa"
This setup assumes the user has the ability to connect to the remote machine using ssh -i /home/user/.ssh/id_rsa user@host.hostname.org and has write-permissions on the remote directory /home/user/.cache/covalent (if it exists) or the closest parent directory (if it does not).
Within a workflow, users can decorate electrons using the default settings:
import covalent as ct
@ct.electron(executor="ssh")
def my_task():
import socket
return socket.gethostname()
or use a class object to customize behavior within particular tasks:
executor = ct.executor.SSHExecutor(
username="user",
hostname="host2.hostname.org",
remote_dir="/tmp/covalent",
ssh_key_file="/home/user/.ssh/host2/id_rsa",
)
@ct.electron(executor=executor)
def my_custom_task(x, y):
return x + y
Dispatcher#
Dispatching jobs to the server and stopping triggered dispatches
Triggers#
Execute a workflow triggered by a specific type of event
Classes:
|
Base class to be subclassed by any custom defined trigger. |
|
Database trigger which can read for changes in a database and trigger workflows based on record changes. |
|
Directory or File based trigger which watches for events in said file/dir and performs a trigger action whenever they happen. |
|
SQLite based Trigger which can read for changes in a SQLite database and trigger workflows based on that. |
|
Performs a trigger action every time_gap seconds. |
- class covalent.triggers.BaseTrigger(lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
objectBase class to be subclassed by any custom defined trigger. Implements all the necessary methods used for interacting with dispatches, including getting their statuses and performing a redispatch of them whenever the trigger gets triggered.
- Parameters
lattice_dispatch_id (
Optional[str]) – Dispatch ID of the worfklow which has to be redispatched in case this trigger gets triggereddispatcher_addr (
Optional[str]) – Address of dispatcher server used to retrieve info about or redispatch any dispatchestriggers_server_addr (
Optional[str]) – Address of the Triggers server (if there is any) to register this trigger to, uses the dispatcher’s address by default
- self.lattice_dispatch_id#
Dispatch ID of the worfklow which has to be redispatched in case this trigger gets triggered
- self.dispatcher_addr#
Address of dispatcher server used to retrieve info about or redispatch any dispatches
- self.triggers_server_addr#
Address of the Triggers server (if there is any) to register this trigger to, uses the dispatcher’s address by default
- self.new_dispatch_ids#
List of all the newly created dispatch ids from performing redispatch
- self.observe_blocks#
Boolean to indicate whether the self.observe method is a blocking call
- self.event_loop#
Event loop to be used if directly calling dispatcher’s functions instead of the REST APIs
- self.use_internal_funcs#
Boolean indicating whether to use dispatcher’s functions directly instead of through API calls
- self.stop_flag#
To handle stopping mechanism in a thread safe manner in case self.observe() is a blocking call (e.g. see TimeTrigger)
Methods:
observe()Start observing for any change which can be used to trigger this trigger.
register()Register this trigger to the Triggers server and start observing.
stop()Stop observing for changes.
to_dict()Return a dictionary representation of this trigger which can later be used to regenerate it.
trigger()Trigger this trigger and perform a redispatch of the connected dispatch id’s workflow.
- abstract observe()[source]#
Start observing for any change which can be used to trigger this trigger. To be implemented by the subclass.
- register()[source]#
Register this trigger to the Triggers server and start observing.
- Return type
None
- class covalent.triggers.DatabaseTrigger(db_path, table_name, poll_interval=1, where_clauses=None, trigger_after_n=1, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerDatabase trigger which can read for changes in a database and trigger workflows based on record changes.
- Parameters
db_path (
str) – Connection string for the databasetable_name (
str) – Name of the table to observepoll_interval (
int) – Time in seconds to wait for before reading the database againwhere_clauses (
Optional[List[str]]) – List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the databasetrigger_after_n (
int) – Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.lattice_dispatch_id (
Optional[str]) – Lattice dispatch id of the workflow to be triggereddispatcher_addr (
Optional[str]) – Address of the dispatcher servertriggers_server_addr (
Optional[str]) – Address of the triggers server
- self.db_path#
Connection string for the database
- self.table_name#
Name of the table to observe
- self.poll_interval#
Time in seconds to wait for before reading the database again
- self.where_clauses#
List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the database
- self.trigger_after_n#
Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action as long as where conditions are met or until stop has being called
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
- class covalent.triggers.DirTrigger(dir_path, event_names, batch_size=1, recursive=False, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerDirectory or File based trigger which watches for events in said file/dir and performs a trigger action whenever they happen.
- Parameters
dir_path – Path to the file/dir which is to be observed for events
event_names – List of event names on which to perform the trigger action. Possible options can be a subset of: [“created”, “deleted”, “modified”, “moved”, “closed”].
batch_size (
int) – The number of changes to wait for before performing the trigger action, default is 1.recursive (
bool) – Whether to recursively watch the directory, default is False.
- self.dir_path#
Path to the file/dir which is to be observed for events
- self.event_names#
List of event names on which to perform the trigger action. Possible options can be a subset of: [“created”, “deleted”, “modified”, “moved”, “closed”]
- self.batch_size#
The number of events to wait for before performing the trigger action, default is 1.
- self.recursive#
Whether to recursively watch the directory, default is False.
- self.n_changes#
Number of events since last trigger action. Whenever self.n_changes == self.batch_size a trigger action happens.
Methods:
Dynamically attaches and overrides the “on_*” methods to the handler depending on which ones are requested by the user.
observe()Start observing the file/dir for any possible events among the ones mentioned in self.event_names.
stop()Stop observing the file or directory for changes.
- attach_methods_to_handler()[source]#
Dynamically attaches and overrides the “on_*” methods to the handler depending on which ones are requested by the user.
- Parameters
event_names – List of event names upon which to perform a trigger action
- Return type
None
- class covalent.triggers.SQLiteTrigger(db_path, table_name, poll_interval=1, where_clauses=None, trigger_after_n=1, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerSQLite based Trigger which can read for changes in a SQLite database and trigger workflows based on that.
- Parameters
db_path (
str) – Absolute path to the database filetable_name (
str) – Name of the table to observepoll_interval (
int) – Time in seconds to wait for before reading the database againwhere_clauses (
Optional[List[str]]) – List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the databasetrigger_after_n (
int) – Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.lattice_dispatch_id (
Optional[str]) – Lattice dispatch id of the workflow to be triggereddispatcher_addr (
Optional[str]) – Address of the dispatcher servertriggers_server_addr (
Optional[str]) – Address of the triggers server
- self.db_path#
Absolute path to the database file
- self.table_name#
Name of the table to observe
- self.poll_interval#
Time in seconds to wait for before reading the database again
- self.where_clauses#
List of “WHERE” conditions, e.g. [“id > 2”, “status = pending”], to check when polling the database
- self.trigger_after_n#
Number of times the event must happen after which the workflow will be triggered. e.g value of 2 means workflow will be triggered once the event has occurred twice.
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action as long as where conditions are met or until stop has being called
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
- class covalent.triggers.TimeTrigger(time_gap, lattice_dispatch_id=None, dispatcher_addr=None, triggers_server_addr=None)[source]#
Bases:
covalent.triggers.base.BaseTriggerPerforms a trigger action every time_gap seconds.
- Parameters
time_gap (
int) – Amount of seconds to wait before doing a trigger action
- self.time_gap#
Amount of seconds to wait before doing a trigger action
- self.stop_flag#
Thread safe flag used to check whether the stop condition has been met
Methods:
observe()Keep performing the trigger action every self.time_gap seconds until stop condition has been met.
stop()Stop the running self.observe() method by setting the self.stop_flag flag.
Cancellation#
Cancel a dispatch using the dispatch_id or multiple tasks within a workflow using the task_ids
- covalent._results_manager.results_manager.cancel(dispatch_id, task_ids=None, dispatcher_addr=None)[source]#
Cancel a running dispatch.
- Parameters
dispatch_id (
str) – The dispatch id of the dispatch to be cancelled.task_ids (
Optional[List[int]]) – Optional, list of task ids to cancel within the workflowdispatcher_addr (
Optional[str]) – Dispatcher server address, if None then defaults to the address set in Covalent’s config.
- Return type
str- Returns
Cancellation response
Results#
Collecting and managing results
- class covalent._results_manager.result.Result(lattice, dispatch_id='')[source]#
Result class to store and perform operations on the result obtained from a dispatch.
- lattice#
“Lattice” object which was dispatched.
- results_dir#
Directory where the result will be stored. It’ll be in the format of “<results_dir>/<dispatch_id>/”.
- dispatch_id#
Dispatch id assigned to this dispatch.
- root_dispatch_id#
Dispatch id of the root lattice in a hierarchy of sublattice workflows.
- status#
Status of the result. It’ll be one of the following: - Result.NEW_OBJ: When it is a new result object. - Result.COMPLETED: When processing of all the nodes has completed successfully. - Result.RUNNING: When some node executions are in process. - Result.FAILED: When one or more node executions have failed. - Result.CANCELLED: When the dispatch was cancelled.
- result#
Final result of the dispatch, i.e whatever the “Lattice” was returning as a function.
- inputs#
Inputs sent to the “Lattice” function for dispatching.
- error#
Error due to which the execution failed.
- Functions:
save_result: Save the result object to the passed results directory or to self.results_dir by default. get_all_node_outputs: Return all the outputs of all the node executions.
Attributes:
Dispatch id of current dispatch.
Encoded final result of current dispatch
End time of processing the dispatch.
Error due to which the dispatch failed.
Inputs sent to the “Lattice” function for dispatching.
“Lattice” object which was dispatched.
Final result of current dispatch.
Results directory used to save this result object.
Dispatch id of the root dispatch
Start time of processing the dispatch.
Status of current dispatch.
Methods:
Return output of every node execution.
Get all the node results.
get_node_result(node_id)Return the result of a particular node.
Post-processing method.
- property dispatch_id: str#
Dispatch id of current dispatch.
- Return type
str
- property encoded_result: covalent._workflow.transportable_object.TransportableObject#
Encoded final result of current dispatch
- Return type
TransportableObject
- property end_time: datetime.datetime#
End time of processing the dispatch.
- Return type
datetime
- property error: str#
Error due to which the dispatch failed.
- Return type
str
- get_all_node_outputs()[source]#
Return output of every node execution.
- Parameters
None –
- Returns
A dictionary containing the output of every node execution.
- Return type
node_outputs
- get_all_node_results()[source]#
Get all the node results.
- Parameters
None –
- Returns
A list of dictionaries containing the result of every node execution.
- Return type
node_results
- get_node_result(node_id)[source]#
Return the result of a particular node.
- Parameters
node_id (
int) – The node id.- Returns
- The result of the node containing below in a dictionary format:
node_id: The node id.
node_name: The name of the node.
start_time: The start time of the node execution.
end_time: The end time of the node execution.
status: The status of the node execution.
output: The output of the node unless error occurred in which case None.
error: The error of the node if occurred else None.
sublattice_result: The result of the sublattice if any.
stdout: The stdout of the node execution.
stderr: The stderr of the node execution.
- Return type
node_result
- property inputs: dict#
Inputs sent to the “Lattice” function for dispatching.
- Return type
dict
- property lattice: covalent._workflow.lattice.Lattice#
“Lattice” object which was dispatched.
- Return type
- post_process()[source]#
Post-processing method. This method was introduced to enable manual client-side postprocessing in case automatic post-processing by the server fails (e.g. insufficient dask worker memory)
- Returns
Post-processed result output
- Return type
Any
- property result: Union[int, float, list, dict]#
Final result of current dispatch.
- Return type
Union[int,float,list,dict]
- property results_dir: str#
Results directory used to save this result object.
- Return type
str
- property root_dispatch_id: str#
Dispatch id of the root dispatch
- Return type
str
- property start_time: datetime.datetime#
Start time of processing the dispatch.
- Return type
datetime
- property status: covalent._shared_files.util_classes.Status#
Status of current dispatch.
- Return type
Status
Covalent CLI Tool#
The command line interface (CLI) tool is used to manage the Covalent server.
Setting Defaults#
Default configuration for covalent can be set by defining the environment variable COVALENT_CONFIG_DIR. By default, config files are stored in ~/.config/covalent/covalent.conf.
Example settings in config file
Note
This is a YAML file, so you can use any YAML syntax.
Tip
Each executor comes with its own configuration parameters that is stored in this same config file config file. For example, for SSH plugin, we have the following settings:
[executors.ssh]
username = "user"
hostname = "host.hostname.org"
remote_dir = "/home/user/.cache/covalent"
ssh_key_file = "/home/user/.ssh/id_rsa"
Generated each time covalent is installed and can be found at ~/.config/covalent/covalent.conf
[sdk]
log_dir = "/Users/he-who-must-not-be-named/.cache/covalent"
log_level = "warning"
enable_logging = "false"
executor_dir = "/Users/voldemort/.config/covalent/executor_plugins"
[dispatcher]
address = "localhost"
port = 48008
cache_dir = "/Users/voldemort/.cache/covalent"
results_dir = "results"
log_dir = "/Users/voldemort/.cache/covalent"
[dask]
cache_dir = "/Users/voldemort/.cache/covalent"
log_dir = "/Users/voldemort/.cache/covalent"
mem_per_worker = "auto"
threads_per_worker = 1
num_workers = 8
scheduler_address = "tcp://127.0.0.1:60690"
dashboard_link = "http://127.0.0.1:8787/status"
process_info = "<DaskCluster name='LocalDaskCluster' parent=80903 started>"
pid = 80924
admin_host = "127.0.0.1"
admin_port = 60682
[workflow_data]
db_path = "/Users/voldemort/.local/share/covalent/workflow_db.sqlite"
storage_type = "local"
base_dir = "/Users/voldemort/.local/share/covalent/workflow_data"
[user_interface]
address = "localhost"
port = 48008
log_dir = "/Users/voldemort/.cache/covalent"
dispatch_db = "/Users/voldemort/.cache/covalent/dispatch_db.sqlite"
[executors.local]
log_stdout = "stdout.log"
log_stderr = "stderr.log"
cache_dir = "/Users/voldemort/.cache/covalent"
[executors.dask]
log_stdout = "stdout.log"
log_stderr = "stderr.log"
cache_dir = "/Users/voldemort/.cache/covalent"